A murine model for the del(GJB6-D13S1830) deletion recapitulating the phenotype of human DFNB1 hearing impairment: generation and functional and histopathological study
- PMID: 38605287
- PMCID: PMC11007912
- DOI: 10.1186/s12864-024-10289-z
A murine model for the del(GJB6-D13S1830) deletion recapitulating the phenotype of human DFNB1 hearing impairment: generation and functional and histopathological study
Abstract
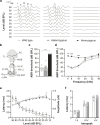
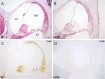
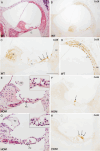
Inherited hearing impairment is a remarkably heterogeneous monogenic condition, involving hundreds of genes, most of them with very small (< 1%) epidemiological contributions. The exception is GJB2, the gene encoding connexin-26 and underlying DFNB1, which is the most frequent type of autosomal recessive non-syndromic hearing impairment (ARNSHI) in most populations (up to 40% of ARNSHI cases). DFNB1 is caused by different types of pathogenic variants in GJB2, but also by large deletions that keep the gene intact but remove an upstream regulatory element that is essential for its expression. Such large deletions, found in most populations, behave as complete loss-of-function variants, usually associated with a profound hearing impairment. By using CRISPR-Cas9 genetic edition, we have generated a murine model (Dfnb1em274) that reproduces the most frequent of those deletions, del(GJB6-D13S1830). Dfnb1em274 homozygous mice are viable, bypassing the embryonic lethality of the Gjb2 knockout, and present a phenotype of profound hearing loss (> 90 dB SPL) that correlates with specific structural abnormalities in the cochlea. We show that Gjb2 expression is nearly abolished and its protein product, Cx26, is nearly absent all throughout the cochlea, unlike previous conditional knockouts in which Gjb2 ablation was not obtained in all cell types. The Dfnb1em274 model recapitulates the clinical presentation of patients harbouring the del(GJB6-D13S1830) variant and thus it is a valuable tool to study the pathological mechanisms of DFNB1 and to assay therapies for this most frequent type of human ARNSHI.
Keywords: GJB2; Connnexin-26; DFNB1; Inherited hearing impairment; Murine models.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Anselmi F, Hernandez VH, Crispino G, Seydel A, Ortolano S, Roper SD, Kessaris N, Richardson W, Rickheit G, Filippov MA, Monyer H, Mammano F. ATP release through connexin hemichannels and gap junction transfer of second messengers propagate Ca2+ signals across the inner ear. Proc Natl Acad Sci U S A. 2008;105(48):18770–18775. doi: 10.1073/pnas.0800793105. - DOI - PMC - PubMed
-
- Behringer R, Gertsenstein M, Vintersten Nagy K, Nagy A. Manipulating the mouse embryo - A laboratory manual. New York: Cold Spring Harbor Laboratory Press; 2014.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

