The integration of single-cell and bulk RNA-seq atlas reveals ERS-mediated acinar cell damage in acute pancreatitis
- PMID: 38605381
- PMCID: PMC11010368
- DOI: 10.1186/s12967-024-05156-0
The integration of single-cell and bulk RNA-seq atlas reveals ERS-mediated acinar cell damage in acute pancreatitis
Abstract
Background: Acute pancreatitis (AP) is a clinically common acute abdominal disease, whose pathogenesis remains unclear. The severe patients usually have multiple complications and lack specific drugs, leading to a high mortality and poor outcome. Acinar cells are recognized as the initial site of AP. However, there are no precise single-cell transcriptomic profiles to decipher the landscape of acinar cells during AP, which are the missing pieces of jigsaw we aimed to complete in this study.
Methods: A single-cell sequencing dataset was used to identify the cell types in pancreas of AP mice and to depict the transcriptomic maps in acinar cells. The pathways' activities were evaluated by gene sets enrichment analysis (GSEA) and single-cell gene sets variation analysis (GSVA). Pseudotime analysis was performed to describe the development trajectories of acinar cells. We also constructed the protein-protein interaction (PPI) network and identified the hub genes. Another independent single-cell sequencing dataset of pancreas samples from AP mice and a bulk RNA sequencing dataset of peripheral blood samples from AP patients were also analyzed.
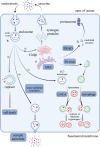
Results: In this study, we identified genetic markers of each cell type in the pancreas of AP mice based on single-cell sequencing datasets and analyzed the transcription changes in acinar cells. We found that acinar cells featured acinar-ductal metaplasia (ADM), as well as increased endocytosis and vesicle transport activity during AP. Notably, the endoplasmic reticulum stress (ERS) and ER-associated degradation (ERAD) pathways activated by accumulation of unfolded/misfolded proteins in acinar cells could be pivotal for the development of AP.
Conclusion: We deciphered the distinct roadmap of acinar cells in the early stage of AP at single-cell level. ERS and ERAD pathways are crucially important for acinar homeostasis and the pathogenesis of AP.
Keywords: Acute pancreatitis; Bulk RNA sequencing; ERAD; ERS; Single-cell RNA sequencing; Vesicle transport.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures








References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

