Development of a customizable mouse backbone spectral flow cytometry panel to delineate immune cell populations in normal and tumor tissues
- PMID: 38605953
- PMCID: PMC11008467
- DOI: 10.3389/fimmu.2024.1374943
Development of a customizable mouse backbone spectral flow cytometry panel to delineate immune cell populations in normal and tumor tissues
Abstract
Introduction: In vivo studies of cancer biology and assessment of therapeutic efficacy are critical to advancing cancer research and ultimately improving patient outcomes. Murine cancer models have proven to be an invaluable tool in pre-clinical studies. In this context, multi-parameter flow cytometry is a powerful method for elucidating the profile of immune cells within the tumor microenvironment and/or play a role in hematological diseases. However, designing an appropriate multi-parameter panel to comprehensively profile the increasing diversity of immune cells across different murine tissues can be extremely challenging.
Methods: To address this issue, we designed a panel with 13 fixed markers that define the major immune populations -referred to as the backbone panel- that can be profiled in different tissues but with the option to incorporate up to seven additional fluorochromes, including any marker specific to the study in question.
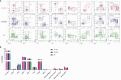
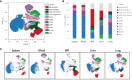
Results: This backbone panel maintains its resolution across different spectral flow cytometers and organs, both hematopoietic and non-hematopoietic, as well as tumors with complex immune microenvironments.
Discussion: Having a robust backbone that can be easily customized with pre-validated drop-in fluorochromes saves time and resources and brings consistency and standardization, making it a versatile solution for immuno-oncology researchers. In addition, the approach presented here can serve as a guide to develop similar types of customizable backbone panels for different research questions requiring high-parameter flow cytometry panels.
Keywords: backbone panel; immune cells; immunophenotyping; mouse; spectral flow cytometry; tumor microenvironment (TME).
Copyright © 2024 Longhini, Fernández-Maestre, Kennedy, Wereski, Mowla, Xiao, Lowe, Levine and Gardner.
Conflict of interest statement
WX has received research support from Stemline Therapeutics. SL serves on the scientific advisory board and has equity in ORIC Pharmaceuticals, Blueprint Medicines, Mirimus Inc, Senecea Therapeutics, Faeth Therapeutics, and PMV Pharmaceuticals. RL is a scientific advisor to Imago, Mission Bio, Zentalis, Ajax, Auron, Prelude, C4 Therapeutics, and Isoplexis, and sits on the supervisory board of Qiagen. RL has also received research support from Ajax, Zentalis, and Abbvie, and has consulted for Incyte, Janssen, Novartis, and AstraZeneca. Additionally, RL has received honoraria from AstraZeneca and Kura for invited lectures, and from Gilead for grant reviews. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

