Deciphering the developmental trajectory of tissue-resident Foxp3+ regulatory T cells
- PMID: 38605970
- PMCID: PMC11007185
- DOI: 10.3389/fimmu.2024.1331846
Deciphering the developmental trajectory of tissue-resident Foxp3+ regulatory T cells
Abstract
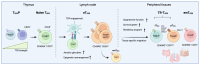
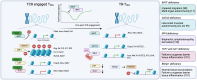
Foxp3+ TREG cells have been at the focus of intense investigation for their recognized roles in preventing autoimmunity, facilitating tissue recuperation following injury, and orchestrating a tolerance to innocuous non-self-antigens. To perform these critical tasks, TREG cells undergo deep epigenetic, transcriptional, and post-transcriptional changes that allow them to adapt to conditions found in tissues both at steady-state and during inflammation. The path leading TREG cells to express these tissue-specialized phenotypes begins during thymic development, and is further driven by epigenetic and transcriptional modifications following TCR engagement and polarizing signals in the periphery. However, this process is highly regulated and requires TREG cells to adopt strategies to avoid losing their regulatory program altogether. Here, we review the origins of tissue-resident TREG cells, from their thymic and peripheral development to the transcriptional regulators involved in their tissue residency program. In addition, we discuss the distinct signalling pathways that engage the inflammatory adaptation of tissue-resident TREG cells, and how they relate to their ability to recognize tissue and pathogen-derived danger signals.
Keywords: Foxp3 + eTREG cells; TREG development; inflammation; mucosal immunity; polarization; tissue residency; transcriptional adaptation.
Copyright © 2024 Alvarez, Liu, Bay and Piccirillo.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

