pH-responsive niosome-based nanocarriers of antineoplastic agents
- PMID: 38606056
- PMCID: PMC11008427
- DOI: 10.1039/d4ra01334d
pH-responsive niosome-based nanocarriers of antineoplastic agents
Abstract
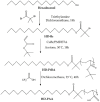
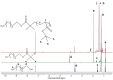
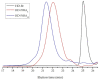
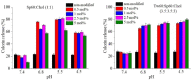
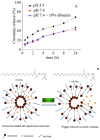
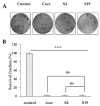
Differences in pH between the tumour interstitium and healthy tissues can be used to induce conformational changes in the nanocarrier structure, thereby triggering drug release at the desired site. In the present study, novel pH-responsive nanocarriers were developed by modifying conventional niosomes with hexadecyl-poly(acrylic acid)n copolymers (HD-PAAn). Niosomal vesicles were prepared by the thin film hydration method using Span 60, Span 60/Tween 60 and cholesterol as main constituents, and HD-PAA modifiers of different concentrations (0.5, 1, 2.5, 5 mol%). Next, two model substances, a water-soluble fluorescent dye (calcein) and a hydrophobic agent with pronounced antineoplastic activity (curcumin), were loaded in the aqueous core and hydrophobic membrane of the elaborated niosomes, respectively. Physicochemical properties of blank and loaded nanocarriers such as hydrodynamic diameter (Dh), size distribution, zeta potential, morphology and pH-responsiveness were investigated in detail. The cytotoxicity of niosomal curcumin was evaluated against human malignant cell lines of different origins (MJ, T-24, HUT-78), and the mechanistic aspects of proapoptotic effects were elucidated. The formulation composed of Span 60/Tween 60/cholesterol/2.5% HD-PAA17 exhibited optimal physicochemical characteristics (Dh 302 nm; ζ potential -22.1 mV; high curcumin entrapment 83%), pH-dependent drug release and improved cytotoxic and apoptogenic activity compared to free curcumin.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures






Similar articles
-
A comparative study on biopharmaceutical function of curcumin and miR-34a by multistimuli-responsive nanoniosome carrier: In-vitro and in-vivo.Front Mol Biosci. 2022 Oct 17;9:1043277. doi: 10.3389/fmolb.2022.1043277. eCollection 2022. Front Mol Biosci. 2022. PMID: 36325275 Free PMC article.
-
Formulation and Evaluation of Hybrid Niosomal In Situ Gel for Intravesical Co-Delivery of Curcumin and Gentamicin Sulfate.Pharmaceutics. 2022 Mar 30;14(4):747. doi: 10.3390/pharmaceutics14040747. Pharmaceutics. 2022. PMID: 35456581 Free PMC article.
-
Green tea catechin loaded niosomes: formulation and their characterization for food fortification.J Food Sci Technol. 2022 Sep;59(9):3669-3682. doi: 10.1007/s13197-022-05384-6. Epub 2022 Mar 15. J Food Sci Technol. 2022. PMID: 35875240 Free PMC article.
-
Preparing and assessing the physiochemical properties of curcumin niosomes and evaluating their cytotoxicity in 3T3 and MCF-7 cell lines.Avicenna J Phytomed. 2021 Jul-Aug;11(4):417-427. doi: 10.22038/AJP.2021.18163. Avicenna J Phytomed. 2021. PMID: 34290972 Free PMC article.
-
Synergistic effect of curcumin-Cu and curcumin-Ag nanoparticle loaded niosome: Enhanced antibacterial and anti-biofilm activities.Bioorg Chem. 2021 Oct;115:105116. doi: 10.1016/j.bioorg.2021.105116. Epub 2021 Jun 24. Bioorg Chem. 2021. PMID: 34333420
Cited by
-
A State-of-the-Art Review on Recent Biomedical Application of Polysaccharide-Based Niosomes as Drug Delivery Systems.Polymers (Basel). 2025 Jun 4;17(11):1566. doi: 10.3390/polym17111566. Polymers (Basel). 2025. PMID: 40508808 Free PMC article. Review.
References
-
- Saraf S. Jain A. Tiwari A. Verma A. Panda P. K. Jain S. K. Advances in liposomal drug delivery to cancer: An overview. J. Drug Delivery Sci. Technol. 2020;56:101549.
LinkOut - more resources
Full Text Sources

