A working model for the formation of Robertsonian chromosomes
- PMID: 38606789
- PMCID: PMC11057876
- DOI: 10.1242/jcs.261912
A working model for the formation of Robertsonian chromosomes
Abstract
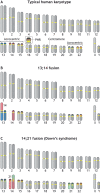
Robertsonian chromosomes form by fusion of two chromosomes that have centromeres located near their ends, known as acrocentric or telocentric chromosomes. This fusion creates a new metacentric chromosome and is a major mechanism of karyotype evolution and speciation. Robertsonian chromosomes are common in nature and were first described in grasshoppers by the zoologist W. R. B. Robertson more than 100 years ago. They have since been observed in many species, including catfish, sheep, butterflies, bats, bovids, rodents and humans, and are the most common chromosomal change in mammals. Robertsonian translocations are particularly rampant in the house mouse, Mus musculus domesticus, where they exhibit meiotic drive and create reproductive isolation. Recent progress has been made in understanding how Robertsonian chromosomes form in the human genome, highlighting some of the fundamental principles of how and why these types of fusion events occur so frequently. Consequences of these fusions include infertility and Down's syndrome. In this Hypothesis, I postulate that the conditions that allow these fusions to form are threefold: (1) sequence homology on non-homologous chromosomes, often in the form of repetitive DNA; (2) recombination initiation during meiosis; and (3) physical proximity of the homologous sequences in three-dimensional space. This Hypothesis highlights the latest progress in understanding human Robertsonian translocations within the context of the broader literature on Robertsonian chromosomes.
Keywords: Chromosomes; Karyotype; Meiosis; Recombination; Robertsonian; Translocation.
© 2024. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures


Similar articles
-
Quantitative analysis of Robertsonian chromosomes inherited by descendants from multiple Rb heterozygotes of Mus m. Domesticus.Front Cell Dev Biol. 2022 Nov 25;10:1050556. doi: 10.3389/fcell.2022.1050556. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36506103 Free PMC article.
-
Recombination suppression by heterozygous Robertsonian chromosomes in the mouse.Genetics. 1993 Mar;133(3):649-67. doi: 10.1093/genetics/133.3.649. Genetics. 1993. PMID: 8454207 Free PMC article.
-
The Robertsonian phenomenon in the house mouse: mutation, meiosis and speciation.Chromosoma. 2014 Dec;123(6):529-44. doi: 10.1007/s00412-014-0477-6. Epub 2014 Jul 23. Chromosoma. 2014. PMID: 25053180 Review.
-
Tracking Chromosomal Origins in the Northern Italy System of Metacentric Races of the House Mouse.Cytogenet Genome Res. 2022;162(4):214-230. doi: 10.1159/000527106. Epub 2022 Dec 1. Cytogenet Genome Res. 2022. PMID: 36455542
-
Role of acrocentric cen-pter satellite DNA in Robertsonian translocation and chromosomal non-disjunction.Mol Biol Med. 1990 Oct;7(5):437-49. Mol Biol Med. 1990. PMID: 2095460 Review.
Cited by
-
Non-canonical DNA in human and other ape telomere-to-telomere genomes.Nucleic Acids Res. 2025 Apr 10;53(7):gkaf298. doi: 10.1093/nar/gkaf298. Nucleic Acids Res. 2025. PMID: 40226919 Free PMC article.
-
Non-canonical DNA in human and other ape telomere-to-telomere genomes.bioRxiv [Preprint]. 2025 Mar 8:2024.09.02.610891. doi: 10.1101/2024.09.02.610891. bioRxiv. 2025. Update in: Nucleic Acids Res. 2025 Apr 10;53(7):gkaf298. doi: 10.1093/nar/gkaf298. PMID: 39713403 Free PMC article. Updated. Preprint.
-
Assessing Human Ribosomal DNA Variation and Its Association With Phenotypic Outcomes.Bioessays. 2025 Apr;47(4):e202400232. doi: 10.1002/bies.202400232. Epub 2025 Jan 20. Bioessays. 2025. PMID: 39834111 Free PMC article. Review.
-
Linear dicentric chromosomes in bacterial natural isolates reveal common constraints for replicon fusion.mBio. 2025 Jun 11;16(6):e0104625. doi: 10.1128/mbio.01046-25. Epub 2025 May 20. mBio. 2025. PMID: 40391973 Free PMC article.
-
Effects of chromosome number reduction on mitotic and meiotic stability in fission yeast.Genome Biol. 2025 Aug 4;26(1):232. doi: 10.1186/s13059-025-03704-5. Genome Biol. 2025. PMID: 40760724 Free PMC article.
References
-
- Altemose, N., Maslan, A., Smith, O. K., Sundararajan, K., Brown, R. R., Mishra, R., Detweiler, A. M., Neff, N., Miga, K. H., Straight, A. F.et al. . (2022). DiMeLo-seq: a long-read, single-molecule method for mapping protein-DNA interactions genome wide. Nat. Methods 19, 711-723. 10.1038/s41592-022-01475-6 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

