The trichome pattern diversity of Cardamine shares genetic mechanisms with Arabidopsis but differs in environmental drivers
- PMID: 38606947
- PMCID: PMC11637488
- DOI: 10.1093/plphys/kiae213
The trichome pattern diversity of Cardamine shares genetic mechanisms with Arabidopsis but differs in environmental drivers
Erratum in
-
Correction to: The trichome pattern diversity of Cardamine shares genetic mechanisms with Arabidopsis but differs in environmental drivers.Plant Physiol. 2024 Dec 2;196(4):3105. doi: 10.1093/plphys/kiae539. Plant Physiol. 2024. PMID: 39418041 Free PMC article. No abstract available.
Abstract
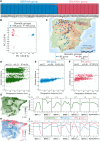
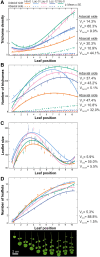
Natural variation in trichome pattern (amount and distribution) is prominent among populations of many angiosperms. However, the degree of parallelism in the genetic mechanisms underlying this diversity and its environmental drivers in different species remain unclear. To address these questions, we analyzed the genomic and environmental bases of leaf trichome pattern diversity in Cardamine hirsuta, a relative of Arabidopsis (Arabidopsis thaliana). We characterized 123 wild accessions for their genomic diversity, leaf trichome patterns at different temperatures, and environmental adjustments. Nucleotide diversities and biogeographical distribution models identified two major genetic lineages with distinct demographic and adaptive histories. Additionally, C. hirsuta showed substantial variation in trichome pattern and plasticity to temperature. Trichome amount in C. hirsuta correlated positively with spring precipitation but negatively with temperature, which is opposite to climatic patterns in A. thaliana. Contrastingly, genetic analysis of C. hirsuta glabrous accessions indicated that, like for A. thaliana, glabrousness is caused by null mutations in ChGLABRA1 (ChGL1). Phenotypic genome-wide association studies (GWAS) further identified a ChGL1 haplogroup associated with low trichome density and ChGL1 expression. Therefore, a ChGL1 series of null and partial loss-of-function alleles accounts for the parallel evolution of leaf trichome pattern in C. hirsuta and A. thaliana. Finally, GWAS also detected other candidate genes (e.g. ChETC3, ChCLE17) that might affect trichome pattern. Accordingly, the evolution of this trait in C. hirsuta and A. thaliana shows partially conserved genetic mechanisms but is likely involved in adaptation to different environments.
© The Author(s) 2024. Published by Oxford University Press on behalf of American Society of Plant Biologists.
Conflict of interest statement
Conflict of interest statement. The authors declare no competing interest.
Figures



Similar articles
-
Cardamine hirsuta: a comparative view.Curr Opin Genet Dev. 2016 Aug;39:1-7. doi: 10.1016/j.gde.2016.05.005. Epub 2016 Jun 5. Curr Opin Genet Dev. 2016. PMID: 27270046 Review.
-
Conservation vs divergence in LEAFY and APETALA1 functions between Arabidopsis thaliana and Cardamine hirsuta.New Phytol. 2017 Oct;216(2):549-561. doi: 10.1111/nph.14419. Epub 2017 Jan 18. New Phytol. 2017. PMID: 28098947
-
Alternate wiring of a KNOXI genetic network underlies differences in leaf development of A. thaliana and C. hirsuta.Genes Dev. 2015 Nov 15;29(22):2391-404. doi: 10.1101/gad.269050.115. Genes Dev. 2015. PMID: 26588991 Free PMC article.
-
The genetic basis for differences in leaf form between Arabidopsis thaliana and its wild relative Cardamine hirsuta.Nat Genet. 2006 Aug;38(8):942-7. doi: 10.1038/ng1835. Epub 2006 Jul 2. Nat Genet. 2006. PMID: 16823378
-
Weeds of change: Cardamine hirsuta as a new model system for studying dissected leaf development.J Plant Res. 2010 Jan;123(1):25-33. doi: 10.1007/s10265-009-0263-3. Epub 2009 Oct 8. J Plant Res. 2010. PMID: 19821009 Review.
Cited by
-
Advancing ecological and evolutionary research in Arabidopsis: Extending insights into model and nonmodel plants.Plant Cell. 2025 Jul 1;37(7):koaf151. doi: 10.1093/plcell/koaf151. Plant Cell. 2025. PMID: 40510015 Free PMC article. Review.
-
Pick a pattern: Transcriptional regulator allelic diversity synergistically drives trichome pattern diversity.Plant Physiol. 2025 Mar 1;197(3):kiaf058. doi: 10.1093/plphys/kiaf058. Plant Physiol. 2025. PMID: 40037573 Free PMC article. No abstract available.
-
The bHLH transcription factor gene EGL3 accounts for the natural diversity in Arabidopsis fruit trichome pattern and morphology.Plant Physiol. 2024 Dec 23;197(1):kiae673. doi: 10.1093/plphys/kiae673. Plant Physiol. 2024. PMID: 39709618 Free PMC article.
References
-
- Arteaga N, Méndez-Vigo B, Fuster-Pons A, Savic M, Murillo-Sánchez A, Picó FX, Alonso-Blanco C. Differential environmental and genomic architectures shape the natural diversity for trichome patterning and morphology in different Arabidopsis organs. Plant Cell Environ. 2022:45(10):3018–3035. 10.1111/pce.14308 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

