A Single-Cell Landscape of Spermioteleosis in Mice and Pigs
- PMID: 38607002
- PMCID: PMC11011153
- DOI: 10.3390/cells13070563
A Single-Cell Landscape of Spermioteleosis in Mice and Pigs
Abstract
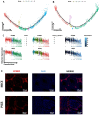
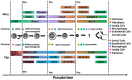
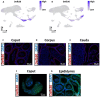
(1) Background: Spermatozoa acquired motility and matured in epididymis after production in the testis. However, there is still limited understanding of the specific characteristics of sperm development across different species. In this study, we employed a comprehensive approach to analyze cell compositions in both testicular and epididymal tissues, providing valuable insights into the changes occurring during meiosis and spermiogenesis in mouse and pig models. Additionally, we identified distinct gene expression signatures associated with various spermatogenic cell types. (2) Methods: To investigate the differences in spermatogenesis between mice and pigs, we constructed a single-cell RNA dataset. (3) Results: Our findings revealed notable differences in testicular cell clusters between these two species. Furthermore, distinct gene expression patterns were observed among epithelial cells from different regions of the epididymis. Interestingly, regional gene expression patterns were also identified within principal cell clusters of the mouse epididymis. Moreover, through analysing differentially expressed genes related to the epididymis in both mouse and pig models, we successfully identified potential marker genes associated with sperm development and maturation for each species studied. (4) Conclusions: This research presented a comprehensive single-cell landscape analysis of both testicular and epididymal tissues, shedding light on the intricate processes involved in spermatogenesis and sperm maturation, specifically within mouse and pig models.
Keywords: mouse; pig; single-cell transcriptome; spermatogenesis.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




Similar articles
-
Single-cell and spatial transcriptomic investigation reveals the spatiotemporal specificity of the beta-defensin gene family during mouse sperm maturation.Cell Commun Signal. 2024 May 14;22(1):267. doi: 10.1186/s12964-024-01637-3. Cell Commun Signal. 2024. PMID: 38745232 Free PMC article.
-
Can we improve time to patency with vasoepididymostomy with an innovative epididymal occlusion stitch?Int Braz J Urol. 2024 Jul-Aug;50(4):504-506. doi: 10.1590/S1677-5538.IBJU.2024.0222. Int Braz J Urol. 2024. PMID: 38743068 Free PMC article.
-
A novel mechanism of sperm midpiece epididymal maturation and the role of CCDC112 in sperm midpiece formation and establishing an optimal flagella waveform.Cell Commun Signal. 2025 Jul 1;23(1):319. doi: 10.1186/s12964-025-02320-x. Cell Commun Signal. 2025. PMID: 40598224 Free PMC article.
-
Improving sperm selection strategies for assisted reproduction through closing the knowledge gap in sperm maturation mechanics.Hum Reprod Open. 2025 Jul 3;2025(3):hoaf040. doi: 10.1093/hropen/hoaf040. eCollection 2025. Hum Reprod Open. 2025. PMID: 40698010 Free PMC article. Review.
-
Gender differences in the context of interventions for improving health literacy in migrants: a qualitative evidence synthesis.Cochrane Database Syst Rev. 2024 Dec 12;12(12):CD013302. doi: 10.1002/14651858.CD013302.pub2. Cochrane Database Syst Rev. 2024. PMID: 39665382
References
-
- Shi J., Fok K.L., Dai P., Qiao F., Zhang M., Liu H., Sang M., Ye M., Liu Y., Zhou Y., et al. Spatio-temporal landscape of mouse epididymal cells and specific mitochondria-rich segments defined by large-scale single-cell RNA-seq. Cell Discov. 2021;7:34. doi: 10.1038/s41421-021-00260-7. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
- 31902158/National Natural Science Foundation of China
- 2021LZGC001/Key Technology R&D Program of Shandong Province
- SDAIT-08-16/Shandong Modern Agricultural Industry Technology System
- K3031222128/Shaanxi Modern Agricultural Industry Technology System
- 2019M662293S/China Postdoctoral Science Foundation
LinkOut - more resources
Full Text Sources

