Cockayne Syndrome Patient iPSC-Derived Brain Organoids and Neurospheres Show Early Transcriptional Dysregulation of Biological Processes Associated with Brain Development and Metabolism
- PMID: 38607030
- PMCID: PMC11011893
- DOI: 10.3390/cells13070591
Cockayne Syndrome Patient iPSC-Derived Brain Organoids and Neurospheres Show Early Transcriptional Dysregulation of Biological Processes Associated with Brain Development and Metabolism
Abstract
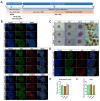
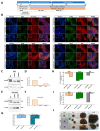
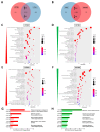
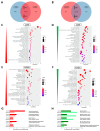
Cockayne syndrome (CS) is a rare hereditary autosomal recessive disorder primarily caused by mutations in Cockayne syndrome protein A (CSA) or B (CSB). While many of the functions of CSB have been at least partially elucidated, little is known about the actual developmental dysregulation in this devasting disorder. Of particular interest is the regulation of cerebral development as the most debilitating symptoms are of neurological nature. We generated neurospheres and cerebral organoids utilizing Cockayne syndrome B protein (CSB)-deficient induced pluripotent stem cells derived from two patients with distinct severity levels of CS and healthy controls. The transcriptome of both developmental timepoints was explored using RNA-Seq and bioinformatic analysis to identify dysregulated biological processes common to both patients with CS in comparison to the control. CSB-deficient neurospheres displayed upregulation of the VEGFA-VEGFR2 signalling pathway, vesicle-mediated transport and head development. CSB-deficient cerebral organoids exhibited downregulation of brain development, neuron projection development and synaptic signalling. We further identified the upregulation of steroid biosynthesis as common to both timepoints, in particular the upregulation of the cholesterol biosynthesis branch. Our results provide insights into the neurodevelopmental dysregulation in patients with CS and strengthen the theory that CS is not only a neurodegenerative but also a neurodevelopmental disorder.
Keywords: Cockayne syndrome; RNA-Seq; brain development; brain organoids; steroid biosynthesis.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures



Similar articles
-
Perspectives in the investigation of Cockayne syndrome group B neurological disease: the utility of patient-derived brain organoid models.J Zhejiang Univ Sci B. 2024 Oct 2;25(10):878-889. doi: 10.1631/jzus.B2300712. J Zhejiang Univ Sci B. 2024. PMID: 39420523 Free PMC article. Review.
-
Multisystem analyses of two Cockayne syndrome associated proteins CSA and CSB reveal shared and unique functions.DNA Repair (Amst). 2019 Nov;83:102696. doi: 10.1016/j.dnarep.2019.102696. Epub 2019 Sep 12. DNA Repair (Amst). 2019. PMID: 31546172
-
Cockayne Syndrome Group B (CSB): The Regulatory Framework Governing the Multifunctional Protein and Its Plausible Role in Cancer.Cells. 2021 Apr 10;10(4):866. doi: 10.3390/cells10040866. Cells. 2021. PMID: 33920220 Free PMC article. Review.
-
Dysregulation of gene expression as a cause of Cockayne syndrome neurological disease.Proc Natl Acad Sci U S A. 2014 Oct 7;111(40):14454-9. doi: 10.1073/pnas.1412569111. Epub 2014 Sep 23. Proc Natl Acad Sci U S A. 2014. PMID: 25249633 Free PMC article.
-
Cockayne syndrome proteins CSA and CSB maintain mitochondrial homeostasis through NAD+ signaling.Aging Cell. 2020 Dec;19(12):e13268. doi: 10.1111/acel.13268. Epub 2020 Nov 9. Aging Cell. 2020. PMID: 33166073 Free PMC article.
Cited by
-
Human iPSC-derived cerebral organoids reveal oxytocin-mediated protection against amyloid-β pathology.Regen Ther. 2025 Jun 26;30:259-267. doi: 10.1016/j.reth.2025.06.013. eCollection 2025 Dec. Regen Ther. 2025. PMID: 40654516 Free PMC article.
-
HiPSC-derived 3D neural models reveal neurodevelopmental pathomechanisms of the Cockayne Syndrome B.Cell Mol Life Sci. 2024 Aug 23;81(1):368. doi: 10.1007/s00018-024-05406-w. Cell Mol Life Sci. 2024. PMID: 39179905 Free PMC article.
-
ATP-Dependent Chromatin Remodeler CSB Couples DNA Repair Pathways to Transcription with Implications for Cockayne Syndrome and Cancer Therapy.Cells. 2025 Feb 7;14(4):239. doi: 10.3390/cells14040239. Cells. 2025. PMID: 39996712 Free PMC article. Review.
-
Case report: Whole exome sequencing identifies a novel variant in the HPRT1 gene in a male with developmental delay.Front Genet. 2025 Feb 28;16:1512070. doi: 10.3389/fgene.2025.1512070. eCollection 2025. Front Genet. 2025. PMID: 40092560 Free PMC article.
-
Cockayne Syndrome Linked to Elevated R-Loops Induced by Stalled RNA Polymerase II during Transcription Elongation.Nat Commun. 2024 Jul 17;15(1):6031. doi: 10.1038/s41467-024-50298-w. Nat Commun. 2024. PMID: 39019869 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

