The ZIKV NS5 Protein Aberrantly Alters the Tubulin Cytoskeleton, Induces the Accumulation of Autophagic p62 and Affects IFN Production: HDAC6 Has Emerged as an Anti-NS5/ZIKV Factor
- PMID: 38607037
- PMCID: PMC11011779
- DOI: 10.3390/cells13070598
The ZIKV NS5 Protein Aberrantly Alters the Tubulin Cytoskeleton, Induces the Accumulation of Autophagic p62 and Affects IFN Production: HDAC6 Has Emerged as an Anti-NS5/ZIKV Factor
Abstract
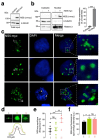
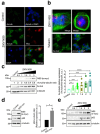
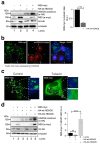
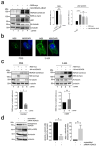
Zika virus (ZIKV) infection and pathogenesis are linked to the disruption of neurogenesis, congenital Zika syndrome and microcephaly by affecting neural progenitor cells. Nonstructural protein 5 (NS5) is the largest product encoded by ZIKV-RNA and is important for replication and immune evasion. Here, we studied the potential effects of NS5 on microtubules (MTs) and autophagy flux, together with the interplay of NS5 with histone deacetylase 6 (HDAC6). Fluorescence microscopy, biochemical cell-fractionation combined with the use of HDAC6 mutants, chemical inhibitors and RNA interference indicated that NS5 accumulates in nuclear structures and strongly promotes the acetylation of MTs that aberrantly reorganize in nested structures. Similarly, NS5 accumulates the p62 protein, an autophagic-flux marker. Therefore, NS5 alters events that are under the control of the autophagic tubulin-deacetylase HDAC6. HDAC6 appears to degrade NS5 by autophagy in a deacetylase- and BUZ domain-dependent manner and to control the cytoplasmic expression of NS5. Moreover, NS5 inhibits RNA-mediated RIG-I interferon (IFN) production, resulting in greater activity when autophagy is inhibited (i.e., effect correlated with NS5 stability). Therefore, it is conceivable that NS5 contributes to cell toxicity and pathogenesis, evading the IFN-immune response by overcoming HDAC6 functions. HDAC6 has emerged as an anti-ZIKV factor by targeting NS5.
Keywords: BUZ domain; HDAC6; MT acetylation; NS5; NS5 clearance; ZIKV; aberrant MTs; autophagic p62; inhibition of IFN production; tubulin deacetylase.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures
Similar articles
-
Cleavage of SQSTM1/p62 by the Zika virus protease NS2B3 prevents autophagic degradation of viral NS3 and NS5 proteins.Autophagy. 2024 Dec;20(12):2769-2784. doi: 10.1080/15548627.2024.2390810. Epub 2024 Aug 17. Autophagy. 2024. PMID: 39128850 Free PMC article.
-
Zika virus-induced fetal demise is triggered by strain- and dose-specific RLR-driven activation of the interferon response in the decidua, placenta, and fetus in Ifnar1-/- mice.J Virol. 2025 Jun 17;99(6):e0066625. doi: 10.1128/jvi.00666-25. Epub 2025 May 22. J Virol. 2025. PMID: 40401980 Free PMC article.
-
Congenital Zika syndrome: Pitfalls in the placental barrier.Rev Med Virol. 2018 Sep;28(5):e1985. doi: 10.1002/rmv.1985. Epub 2018 May 15. Rev Med Virol. 2018. PMID: 29761581
-
African strains of Zika virus resist ISG-mediated restriction.PLoS Negl Trop Dis. 2025 Jul 14;19(7):e0013326. doi: 10.1371/journal.pntd.0013326. eCollection 2025 Jul. PLoS Negl Trop Dis. 2025. PMID: 40658726 Free PMC article.
-
Chemical Versatility in Catalysis and Inhibition of the Class IIb Histone Deacetylases.Acc Chem Res. 2024 Apr 16;57(8):1135-1148. doi: 10.1021/acs.accounts.3c00801. Epub 2024 Mar 26. Acc Chem Res. 2024. PMID: 38530703 Free PMC article. Review.
Cited by
-
The function and mechanism of protein acylation in the regulation of viral infection.Virulence. 2025 Dec;16(1):2530171. doi: 10.1080/21505594.2025.2530171. Epub 2025 Jul 17. Virulence. 2025. PMID: 40673681 Free PMC article. Review.
-
Acetylation in Viral Infection and Disease.Results Probl Cell Differ. 2025;75:329-361. doi: 10.1007/978-3-031-91459-1_12. Results Probl Cell Differ. 2025. PMID: 40593216 Review.
-
Emerging Arthropod-Borne Viruses Hijack the Host Cell Cytoskeleton During Neuroinvasion.Viruses. 2025 Jun 26;17(7):908. doi: 10.3390/v17070908. Viruses. 2025. PMID: 40733526 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
- UNLL10-3E-783/European Regional Development Fund (ERDF) and "Fundación CajaCanarias"
- PID2021-123031OB-I00/MCIN ("Ministerio de Ciencia e Innovación")/AEI ("Agencia Española de Investigación")/10.13039/501100011033/FEDER, UE) (Spain)
- SEGAI-ULL/SEGAI-ULL
- ProID2020010093/Agencia Canaria de Investigación, Innovación y Sociedad de la Información" and European Social Fund
- Contrato Predoctoral Ministerio-ULL Formación de Doctores" (2019 Program)/Contrato Predoctoral Ministerio-ULL Formación de Doctores" (2019 Program)
LinkOut - more resources
Full Text Sources
Medical
Research Materials

