Discovery of First-in-Class PROTAC Degraders of SARS-CoV-2 Main Protease
- PMID: 38608245
- PMCID: PMC11056980
- DOI: 10.1021/acs.jmedchem.3c02416
Discovery of First-in-Class PROTAC Degraders of SARS-CoV-2 Main Protease
Abstract
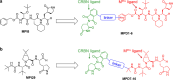
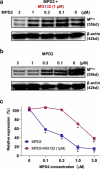
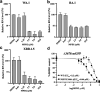
We have witnessed three coronavirus (CoV) outbreaks in the past two decades, including the COVID-19 pandemic caused by SARS-CoV-2. Main protease (MPro), a highly conserved protease among various CoVs, is essential for viral replication and pathogenesis, making it a prime target for antiviral drug development. Here, we leverage proteolysis targeting chimera (PROTAC) technology to develop a new class of small-molecule antivirals that induce the degradation of SARS-CoV-2 MPro. Among them, MPD2 was demonstrated to effectively reduce MPro protein levels in 293T cells, relying on a time-dependent, CRBN-mediated, and proteasome-driven mechanism. Furthermore, MPD2 exhibited remarkable efficacy in diminishing MPro protein levels in SARS-CoV-2-infected A549-ACE2 cells. MPD2 also displayed potent antiviral activity against various SARS-CoV-2 strains and exhibited enhanced potency against nirmatrelvir-resistant viruses. Overall, this proof-of-concept study highlights the potential of targeted protein degradation of MPro as an innovative approach for developing antivirals that could fight against drug-resistant viral variants.
Conflict of interest statement
The authors declare no competing financial interest.
Figures






Update of
-
Discovery of First-in-Class PROTAC Degraders of SARS-CoV-2 Main Protease.bioRxiv [Preprint]. 2023 Sep 29:2023.09.29.560163. doi: 10.1101/2023.09.29.560163. bioRxiv. 2023. Update in: J Med Chem. 2024 Apr 25;67(8):6495-6507. doi: 10.1021/acs.jmedchem.3c02416. PMID: 37808777 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

