An On-Farm Workflow for Predictive Management of Paralytic Shellfish Toxin-Producing Harmful Algal Blooms for the Aquaculture Industry
- PMID: 38608723
- PMCID: PMC11044886
- DOI: 10.1021/acs.est.3c10502
An On-Farm Workflow for Predictive Management of Paralytic Shellfish Toxin-Producing Harmful Algal Blooms for the Aquaculture Industry
Abstract
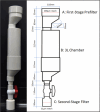
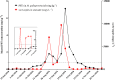
Paralytic shellfish toxins (PSTs) produced by marine dinoflagellates significantly impact shellfish industries worldwide. Early detection on-farm and with minimal training would allow additional time for management decisions to minimize economic losses. Here, we describe and test a standardized workflow based on the detection of sxtA4, an initial gene in the biosynthesis of PSTs. The workflow is simple and inexpensive and does not require a specialized laboratory. It consists of (1) water collection and filtration using a custom gravity sampler, (2) buffer selection for sample preservation and cell lysis for DNA, and (3) an assay based on a region of sxtA, DinoDtec lyophilized quantitative polymerase chain reaction (qPCR) assay. Water samples spiked with Alexandrium catenella showed a cell recovery of >90% when compared to light microscopy counts. The performance of the lysis method (90.3% efficient), Longmire's buffer, and the DinoDtec qPCR assay (tested across a range of Alexandrium species (90.7-106.9% efficiency; r2 > 0.99)) was found to be specific, sensitive, and efficient. We tested the application of this workflow weekly from May 2016 to 30th October 2017 to compare the relationship between sxtA4 copies L-1 in seawater and PSTs in mussel tissue (Mytilus galloprovincialis) on-farm and spatially (across multiple sites), effectively demonstrating an ∼2 week early warning of two A. catenella HABs (r = 0.95). Our tool provides an early, accurate, and efficient method for the identification of PST risk in shellfish aquaculture.
Keywords: alexandrium spp.; aquaculture industry; harmful algal blooms (HABs); molecular detection; on-farm workflow; paralytic shellfish toxins (PSTs); sxtA4 gene.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


References
-
- Anderson D. M. Bloom dynamics of toxic Alexandrium species in the northeastern U.S. Limnol. Oceanogr. 1997, 42 (2), 1009–1022. 10.4319/lo.1997.42.5_part_2.1009. - DOI
-
- Chapelle A.; Le Gac M.; Labry C.; Siano R.; Quere J.; Caradec F.. The Bay of Brest (France), A New Risky Site for Toxic Alexandrium Minutum Blooms and PSP Shellfish Contamination; Reguera B.; Bresnan E., Eds.; Harmful Algae News, 2015; Vol. 51, pp 4–5.
-
- Hallegraeff G. M.; Bolch C. Unprecedented toxic algal blooms impact on Tasmanian seafood industry. Microbiol. Aust. 2016, 37 (3), 143–144. 10.1071/MA16049. - DOI
-
- Usup G.; Pin L. C.; Ahmad A.; Teen L. P. Phylogenetic relationship of Alexandrium tamiyavanichii (Dinophyceae) to other Alexandrium species based on ribosomal RNA gene sequences. Harmful Algae 2002, 1 (1), 59–68. 10.1016/S1568-9883(02)00003-3. - DOI
-
- Campbell A.; Hudson D.; McLeod C.; Nicholls C.; Pointon A.. Tactical Research Fund: Review of the 2012 Paralytic Shellfish Toxin Event in Tasmania Associated with the Dinoflagellate Alga, Alexandrium tamarense Safe Fish: Adelaide; 2013.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

