Disentangling the riddle of systemic lupus erythematosus with antiphospholipid syndrome: blood transcriptome analysis reveals a less-pronounced IFN-signature and distinct molecular profiles in venous versus arterial events
- PMID: 38609158
- PMCID: PMC11420729
- DOI: 10.1136/ard-2024-225664
Disentangling the riddle of systemic lupus erythematosus with antiphospholipid syndrome: blood transcriptome analysis reveals a less-pronounced IFN-signature and distinct molecular profiles in venous versus arterial events
Abstract
Introduction: Systemic lupus erythematosus with antiphospholipid syndrome (SLE-APS) represents a challenging SLE endotype whose molecular basis remains unknown.
Methods: We analysed whole-blood RNA-sequencing data from 299 patients with SLE (108 SLE-antiphospholipid antibodies (aPL)-positive, including 67 SLE-APS; 191 SLE-aPL-negative) and 72 matched healthy controls (HC). Pathway enrichment analysis, unsupervised weighted gene coexpression network analysis and machine learning were applied to distinguish disease endotypes.
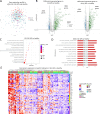
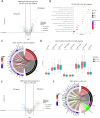
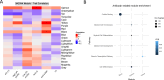
Results: Patients with SLE-APS demonstrated upregulated type I and II interferon (IFN) pathways compared with HC. Using a 100-gene random forests model, we achieved a cross-validated accuracy of 75.6% in distinguishing these two states. Additionally, the comparison between SLE-APS and SLE-aPL-negative revealed 227 differentially expressed genes, indicating downregulation of IFN-α and IFN-γ signatures, coupled with dysregulation of the complement cascade, B-cell activation and neutrophil degranulation. Unsupervised analysis of SLE transcriptome identified 21 gene modules, with SLE-APS strongly linked to upregulation of the 'neutrophilic/myeloid' module. Within SLE-APS, venous thromboses positively correlated with 'neutrophilic/myeloid' and 'B cell' modules, while arterial thromboses were associated with dysregulation of 'DNA damage response (DDR)' and 'metabolism' modules. Anticardiolipin and anti-β2GPI positivity-irrespective of APS status-were associated with the 'neutrophilic/myeloid' and 'protein-binding' module, respectively.
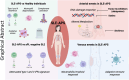
Conclusions: There is a hierarchical upregulation and-likely-dependence on IFN in SLE with the highest IFN signature observed in SLE-aPL-negative patients. Venous thrombotic events are associated with neutrophils and B cells while arterial events with DDR and impaired metabolism. This may account for their differential requirements for anticoagulation and provide rationale for the potential use of mTOR inhibitors such as sirolimus and the direct fIIa inhibitor dabigatran in SLE-APS.
Keywords: Antibodies, Antiphospholipid; Antiphospholipid Syndrome; Lupus Erythematosus, Systemic; Thrombosis.
© Author(s) (or their employer(s)) 2024. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ on behalf of EULAR.
Conflict of interest statement
Competing interests: None declared.
Figures


References
-
- Nikolopoulos D, Kostopoulou M, Pieta A, et al. . Evolving phenotype of systemic lupus erythematosus in caucasians: low incidence of lupus nephritis, high burden of neuropsychiatric disease and increased rates of late-onset lupus in the ‘ attikon ’ cohort. Lupus 2020;29:514–22. 10.1177/0961203320908932 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

