Utility of polygenic scores across diverse diseases in a hospital cohort for predictive modeling
- PMID: 38609356
- PMCID: PMC11014845
- DOI: 10.1038/s41467-024-47472-5
Utility of polygenic scores across diverse diseases in a hospital cohort for predictive modeling
Abstract
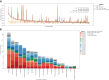
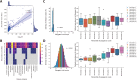
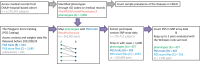
Polygenic scores estimate genetic susceptibility to diseases. We systematically calculated polygenic scores across 457 phenotypes using genotyping array data from China Medical University Hospital. Logistic regression models assessed polygenic scores' ability to predict disease traits. The polygenic score model with the highest accuracy, based on maximal area under the receiver operating characteristic curve (AUC), is provided on the GeneAnaBase website of the hospital. Our findings indicate 49 phenotypes with AUC greater than 0.6, predominantly linked to endocrine and metabolic diseases. Notably, hyperplasia of the prostate exhibited the highest disease prediction ability (P value = 1.01 × 10-19, AUC = 0.874), highlighting the potential of these polygenic scores in preventive medicine and diagnosis. This study offers a comprehensive evaluation of polygenic scores performance across diverse human traits, identifying promising applications for precision medicine and personalized healthcare, thereby inspiring further research and development in this field.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests. All authors declare that they have no known competing financial interests or non-financial interests that could have appeared to influence the work reported in this paper.
Figures



Similar articles
-
Polygenic risk scores for the prediction of common cancers in East Asians: A population-based prospective cohort study.Elife. 2023 Mar 27;12:e82608. doi: 10.7554/eLife.82608. Elife. 2023. PMID: 36971353 Free PMC article.
-
Association of Genome-Wide Polygenic Scores for Multiple Psychiatric and Common Traits in Preadolescent Youths at Risk of Suicide.JAMA Netw Open. 2022 Feb 1;5(2):e2148585. doi: 10.1001/jamanetworkopen.2021.48585. JAMA Netw Open. 2022. PMID: 35188556 Free PMC article.
-
Incorporating Polygenic Risk Scores and Nongenetic Risk Factors for Breast Cancer Risk Prediction Among Asian Women.JAMA Netw Open. 2022 Mar 1;5(3):e2149030. doi: 10.1001/jamanetworkopen.2021.49030. JAMA Netw Open. 2022. PMID: 35311964 Free PMC article.
-
Systematic Review of Polygenic Risk Scores for Type 1 and Type 2 Diabetes.Int J Mol Sci. 2020 Mar 2;21(5):1703. doi: 10.3390/ijms21051703. Int J Mol Sci. 2020. PMID: 32131491 Free PMC article.
-
Current Developments of Clinical Sequencing and the Clinical Utility of Polygenic Risk Scores in Inflammatory Diseases.Front Immunol. 2021 Jan 29;11:577677. doi: 10.3389/fimmu.2020.577677. eCollection 2020. Front Immunol. 2021. PMID: 33633722 Free PMC article. Review.
Cited by
-
Prediction of risk for isolated incomplete lateral meniscal injury using a dynamic nomogram based on MRI-derived anatomic radiomics and physical activity: a proof-of-concept study in 3PM-guided management.EPMA J. 2025 Jan 27;16(1):199-215. doi: 10.1007/s13167-025-00399-3. eCollection 2025 Mar. EPMA J. 2025. PMID: 39991097
-
Diversity and longitudinal records: Genetic architecture of disease associations and polygenic risk in the Taiwanese Han population.Sci Adv. 2025 Jun 6;11(23):eadt0539. doi: 10.1126/sciadv.adt0539. Epub 2025 Jun 4. Sci Adv. 2025. PMID: 40465716 Free PMC article.
-
Predictive capabilities of polygenic scores in an East-Asian population-based cohort: the Singapore Chinese health study.Commun Biol. 2025 Aug 15;8(1):1228. doi: 10.1038/s42003-025-08675-8. Commun Biol. 2025. PMID: 40817126 Free PMC article.
-
Discovery and prioritization of genetic determinants of kidney function in 297,355 individuals from Taiwan and Japan.Nat Commun. 2024 Oct 29;15(1):9317. doi: 10.1038/s41467-024-53516-7. Nat Commun. 2024. PMID: 39472450 Free PMC article.
References
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

