Predicting permeation of compounds across the outer membrane of P. aeruginosa using molecular descriptors
- PMID: 38609430
- PMCID: PMC11015012
- DOI: 10.1038/s42004-024-01161-y
Predicting permeation of compounds across the outer membrane of P. aeruginosa using molecular descriptors
Abstract
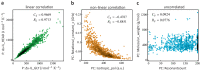
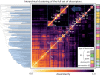
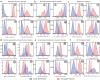
The ability Gram-negative pathogens have at adapting and protecting themselves against antibiotics has increasingly become a public health threat. Data-driven models identifying molecular properties that correlate with outer membrane (OM) permeation and growth inhibition while avoiding efflux could guide the discovery of novel classes of antibiotics. Here we evaluate 174 molecular descriptors in 1260 antimicrobial compounds and study their correlations with antibacterial activity in Gram-negative Pseudomonas aeruginosa. The descriptors are derived from traditional approaches quantifying the compounds' intrinsic physicochemical properties, together with, bacterium-specific from ensemble docking of compounds targeting specific MexB binding pockets, and all-atom molecular dynamics simulations in different subregions of the OM model. Using these descriptors and the measured inhibitory concentrations, we design a statistical protocol to identify predictors of OM permeation/inhibition. We find consistent rules across most of our data highlighting the role of the interaction between the compounds and the OM. An implementation of the rules uncovered in our study is shown, and it demonstrates the accuracy of our approach in a set of previously unseen compounds. Our analysis sheds new light on the key properties drug candidates need to effectively permeate/inhibit P. aeruginosa, and opens the gate to similar data-driven studies in other Gram-negative pathogens.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
Predictive Rules of Efflux Inhibition and Avoidance in Pseudomonas aeruginosa.mBio. 2021 Jan 19;12(1):e02785-20. doi: 10.1128/mBio.02785-20. mBio. 2021. PMID: 33468691 Free PMC article.
-
Synergy between Active Efflux and Outer Membrane Diffusion Defines Rules of Antibiotic Permeation into Gram-Negative Bacteria.mBio. 2017 Oct 31;8(5):e01172-17. doi: 10.1128/mBio.01172-17. mBio. 2017. PMID: 29089426 Free PMC article.
-
Identification of Efflux Substrates Using a Riboswitch-Based Reporter in Pseudomonas aeruginosa.mSphere. 2023 Apr 20;8(2):e0006923. doi: 10.1128/msphere.00069-23. Epub 2023 Mar 22. mSphere. 2023. PMID: 36946743 Free PMC article.
-
Structure- and Ligand-Dynamics-Based Design of Novel Antibiotics Targeting Lipid A Enzymes LpxC and LpxH in Gram-Negative Bacteria.Acc Chem Res. 2021 Apr 6;54(7):1623-1634. doi: 10.1021/acs.accounts.0c00880. Epub 2021 Mar 15. Acc Chem Res. 2021. PMID: 33720682 Free PMC article. Review.
-
Synergy by Perturbing the Gram-Negative Outer Membrane: Opening the Door for Gram-Positive Specific Antibiotics.ACS Infect Dis. 2022 Sep 9;8(9):1731-1757. doi: 10.1021/acsinfecdis.2c00193. Epub 2022 Aug 10. ACS Infect Dis. 2022. PMID: 35946799 Free PMC article. Review.
Cited by
-
[1,3]Thiazolo[3,2-b][1,2,4]triazolium Salts as Effective Antimicrobial Agents: Synthesis, Biological Activity Evaluation, and Molecular Docking Studies.Int J Mol Sci. 2025 Jul 16;26(14):6845. doi: 10.3390/ijms26146845. Int J Mol Sci. 2025. PMID: 40725092 Free PMC article.
References
-
- World Health Organization. Antibacterial agents in clinical development: An analysis of the antibacterial clinical development pipeline, including tuberculosis. Tech. Rep., (World Health Organization, 2017). http://www.jstor.org/stable/resrep35853.1.
Grants and funding
LinkOut - more resources
Full Text Sources

