Evaluation of EGFR and COX pathway inhibition in human colon organoids of serrated polyposis and other hereditary cancer syndromes
- PMID: 38609520
- PMCID: PMC11512843
- DOI: 10.1007/s10689-024-00370-7
Evaluation of EGFR and COX pathway inhibition in human colon organoids of serrated polyposis and other hereditary cancer syndromes
Abstract
Serrated polyposis syndrome (SPS) presents with multiple sessile serrated lesions (SSL) in the large intestine and confers increased colorectal cancer (CRC) risk. However, the etiology of SPS is not known. SSL-derived organoids have not been previously studied but may help provide insights into SPS pathogenesis and identify novel biomarkers and chemopreventive strategies. This study examined effects of EGFR and COX pathway inhibition in organoid cultures derived from uninvolved colon and polyps of SPS patients. We also compared with organoids representing the hereditary gastrointestinal syndromes, Familial Adenomatous Polyposis (FAP) and Lynch syndrome (LS). Eighteen total organoid colon cultures were generated from uninvolved colon and polyps in SPS, FAP, LS, and non-syndromic screening colonoscopy patients. BRAF and KRAS mutation status was determined for each culture. Erlotinib (EGFR inhibitor) and sulindac (COX inhibitor) were applied individually and in combination. A 44-target gene custom mRNA panel (including WNT and COX pathway genes) and a 798-gene microRNA gene panel were used to quantitate organoid RNA expression by NanoString analysis. Erlotinib treatment significantly decreased levels of mRNAs associated with WNT and MAPK kinase signaling in organoids from uninvolved colon from all four patient categories and from all SSL and adenomatous polyps. Sulindac did not change the mRNA profile in any culture. Our findings suggest that EGFR inhibitors may contribute to the chemopreventive treatment of SSLs. These findings may also facilitate clinical trial design using these agents in SPS patients. Differentially expressed genes identified in our study (MYC, FOSL1, EGR1, IL33, LGR5 and FOXQ1) may be used to identify other new molecular targets for chemoprevention of SSLs.
Keywords: Colon organoid; Colon polyp; Erlotinib; Familial adenomatous polyposis; Gene expression; Lynch syndrome; Serrated polyposis; Sulindac.
© 2024. The Author(s).
Conflict of interest statement
Patent - Methods and compositions for predicting a colon cancer subtype Inventors - Don Delker, Priyanka Kanth Publication date - 2022/3/22 Patent office - US Patent number - 11279980
None.
Figures

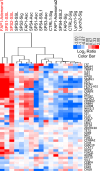
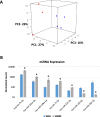


References
-
- National Center for Health Statistics. Health, United States, 2010: With Special Feature on Death and Dying. (U.S. Government) (2011) - PubMed
-
- Kanth P, Grimmett J, Champine M, Burt R, Samadder NJ (2017) Hereditary colorectal polyposis and cancer syndromes: a primer on diagnosis and management. Am J Gastroenterol 112(10):1509–1525 - PubMed
-
- Schmelz EM, Xu H, Sengupta R et al (2007) Regression of early and intermediate stages of colon cancer by targeting multiple members of the EGFR family with EGFR-related protein. Cancer Res 67(11):5389–5396 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

