Blimp-1 and c-Maf regulate immune gene networks to protect against distinct pathways of pathobiont-induced colitis
- PMID: 38609547
- PMCID: PMC11065689
- DOI: 10.1038/s41590-024-01814-z
Blimp-1 and c-Maf regulate immune gene networks to protect against distinct pathways of pathobiont-induced colitis
Abstract
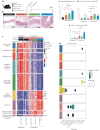
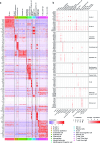
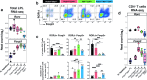
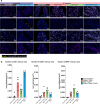
Intestinal immune responses to microbes are controlled by the cytokine IL-10 to avoid immune pathology. Here, we use single-cell RNA sequencing of colon lamina propria leukocytes (LPLs) along with RNA-seq and ATAC-seq of purified CD4+ T cells to show that the transcription factors Blimp-1 (encoded by Prdm1) and c-Maf co-dominantly regulate Il10 while negatively regulating proinflammatory cytokines in effector T cells. Double-deficient Prdm1fl/flMaffl/flCd4Cre mice infected with Helicobacter hepaticus developed severe colitis with an increase in TH1/NK/ILC1 effector genes in LPLs, while Prdm1fl/flCd4Cre and Maffl/flCd4Cre mice exhibited moderate pathology and a less-marked type 1 effector response. LPLs from infected Maffl/flCd4Cre mice had increased type 17 responses with increased Il17a and Il22 expression and an increase in granulocytes and myeloid cell numbers, resulting in increased T cell-myeloid-neutrophil interactions. Genes over-expressed in human inflammatory bowel disease showed differential expression in LPLs from infected mice in the absence of Prdm1 or Maf, revealing potential mechanisms of human disease.
© 2024. The Author(s).
Conflict of interest statement
F.P. received grant funding or consultancy fees from Roche, Genentech, GSK, Janssen, Novartis and T Cypher. The other authors declare no competing interests.
Figures












References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

