Identification of hub genes and diagnostic efficacy for triple-negative breast cancer through WGCNA and Mendelian randomization
- PMID: 38609711
- PMCID: PMC11014828
- DOI: 10.1007/s12672-024-00970-w
Identification of hub genes and diagnostic efficacy for triple-negative breast cancer through WGCNA and Mendelian randomization
Abstract
Objective: Triple-negative breast cancer (TNBC) represents a particularly aggressive form of breast cancer with a poor prognosis due to a lack of targeted treatments resulting from limited a understanding of the underlying mechanisms. The aim of this study was the identification of hub genes for TNBC and assess their clinical applicability in predicting the disease.
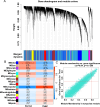
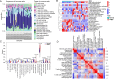
Methods: This study employed a combination of weighted gene co-expression network analysis (WGCNA) and differentially expressed genes (DEGs) to identify new susceptible modules and central genes in TNBC. The potential functional roles of the central genes were investigated using Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) analyses. Furthermore, a predictive model and ROC curve were developed to assess the diagnostic performance of the identified central genes. The correlation between CCNB1 and immune cells proportion was also investigated. At last, a Mendelian randomization (MR) analysis utilizing Genome-Wide Association Study (GWAS) data was analyzed to establish the causal effect of CCNB1 level on TNBC.
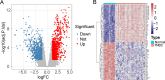
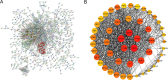
Results: WGCNA was applied to determine gene co-expression maps and identify the most relevant module. Through a screening process, 1585 candidate hub genes were subsequently identified with WGCNA and DEGs. GO and KEGG function enrichment analysis indicated that these core genes were related to various biological processes, such as organelle fission, chromosome segregation, nuclear division, mitotic cell cycle phase transition, the cell cycle, amyotrophic lateral sclerosis, and motor proteins. Using STRING and Cytoscape, the top five genes with high degrees were identified as CDC2, CCNB1, CCNA2, TOP2A, and CCNB2. The nomogram model demonstrated good performance in predicting TNBC risk and was proven effective in diagnosis, as evidenced by the receiver operating characteristic (ROC) curve. Further investigation revealed a causal association between CCNB1 and immune cell infiltrates in TNBC. Survival analysis revealed high expression of the CCNB1 gene leads to poorer prognosis in TNBC patients. Additionally, analysis using inverse variance weighting revealed that CCNB1 was linked to a 2.8% higher risk of TNBC (OR: 1.028, 95% CI 1.002-1.055, p = 0.032).
Conclusion: We established a co-expression network using the WGCNA methodology to detect pivotal genes associated with TNBC. This finding holds promise for advancing the creation of pre-symptomatic diagnostic tools and deepening our comprehension of the pathogenic mechanisms involved in TNBC risk genes.
Keywords: Biomarker; Breast cancer; CCNB1; Mendelian randomization; TNBC; Triple-negative breast cancer; WGCNA.
© 2024. The Author(s).
Conflict of interest statement
The authors have no relevant financial or non-financial interests to disclose.
Figures




References
-
- Garrido-Castro AC, Lin NU, Polyak K. Insights into molecular classifications of triple-negative breast cancer: improving patient selection for treatment. Cancer Discov. 2019;9(2):176–198. doi: 10.1158/2159-8290.CD-18-1177. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
