Evaluation of heterologous expression in Pichia pastoris of Pine Weevil TRPA1 by GFP and flow cytometry
- PMID: 38609906
- PMCID: PMC11015645
- DOI: 10.1186/s12934-024-02382-5
Evaluation of heterologous expression in Pichia pastoris of Pine Weevil TRPA1 by GFP and flow cytometry
Abstract
Background: The wasabi receptor, also known as the Transient Receptor Potential Ankyrin 1 (TRPA1) ion channel, is a potential target for development of repellents for insects, like the pine weevil (Hylobius abietis) feeding on conifer seedlings and causing damage in forestry. Heterologous expression of TRPA1 from pine weevil in the yeast Pichia pastoris can potentially provide protein for structural and functional studies. Here we take advantage of the Green Fluorescent Protein (GFP) tag to examine the various steps of heterologous expression, to get more insight in clone selection, expression and isolation of the intact purified protein.
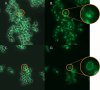
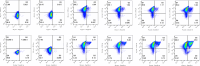
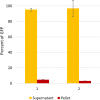
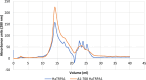
Results: The sequence of HaTRPA1 is reported and GFP-tagged constructs were made of the full-length protein and a truncated version (Δ1-708 HaTRPA1), lacking the N-terminal ankyrin repeat domain. Clones were screened on GFP expression plates, induced in small liquid cultures and in fed-batch cultures, and evaluated by flow cytometry and fluorescence microscopy. The screening on plates successfully identifies low-expression clones, but fails to predict the ranking of the best performing clones in small-scale liquid cultures. The two constructs differ in their cellular localization. Δ1-708 HaTRPA1 is found in a ring at the perimeter of cell, whereas HaTRPA1 is forming highly fluorescent speckles in interior regions of the cell. The pattern is consistent in different clones of the same construct and persists in fed-batch culture. The expression of Δ1-708 HaTRPA1 decreases the viability more than HaTRPA1, and in fed-batch culture it is clear that intact cells first express Δ1-708 HaTRPA1 and then become damaged. Purifications show that both constructs suffer from degradation of the expressed protein, but especially the HaTRPA1 construct.
Conclusions: The GFP tag makes it possible to follow expression by flow cytometry and fluorescence microscopy. Analyses of localization, cell viability and expression show that the former two parameters are specific for each of the two evaluated constructs, whereas the relative expression of the constructs varies with the cultivation method. High expression is not all that matters, so taking damaged cells into account, something that may be linked to protein degradation, is important when picking the most suitable construct, clone, and expression scheme.
Keywords: Hylobius abietis; Ankyrin repeat domain; Flow cytometry; Green Fluorescent Protein; Membrane protein; TRPA1; Viability.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Day BLKR, Damage . Control and Management of Weevil Pests, especially Hylobius Abietis. In: François Lieutier KRD, Battisti A, Grégoire J-C, Evans HF, editors. Bark and Wood Boring insects in living trees in Europe. A Synthesis: Springer; 2004. pp. 414–44.
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources

