Targeting of REST with rationally-designed small molecule compounds exhibits synergetic therapeutic potential in human glioblastoma cells
- PMID: 38609948
- PMCID: PMC11015551
- DOI: 10.1186/s12915-024-01879-0
Targeting of REST with rationally-designed small molecule compounds exhibits synergetic therapeutic potential in human glioblastoma cells
Abstract
Background: Glioblastoma (GBM) is an aggressive brain cancer associated with poor prognosis, intrinsic heterogeneity, plasticity, and therapy resistance. In some GBMs, cell proliferation is fueled by a transcriptional regulator, repressor element-1 silencing transcription factor (REST).
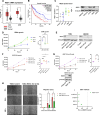
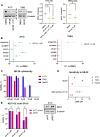
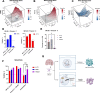
Results: Using CRISPR/Cas9, we identified GBM cell lines dependent on REST activity. We developed new small molecule inhibitory compounds targeting small C-terminal domain phosphatase 1 (SCP1) to reduce REST protein level and transcriptional activity in glioblastoma cells. Top leads of the series like GR-28 exhibit potent cytotoxicity, reduce REST protein level, and suppress its transcriptional activity. Upon the loss of REST protein, GBM cells can potentially compensate by rewiring fatty acid metabolism, enabling continued proliferation. Combining REST inhibition with the blockade of this compensatory adaptation using long-chain acyl-CoA synthetase inhibitor Triacsin C demonstrated substantial synergetic potential without inducing hepatotoxicity.
Conclusions: Our results highlight the efficacy and selectivity of targeting REST alone or in combination as a therapeutic strategy to combat high-REST GBM.
Keywords: Glioblastoma (GBM); Repressor element-1 silencing transcription factor; SCP1 phosphatase; Synergy.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no conflicts of interest related to this research. However, it should be noted that SBP, DS, and YJZ are listed as inventors on a provisional patent related to the technology discussed in this paper.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

