Nuclear lamina component KAKU4 regulates chromatin states and transcriptional regulation in the Arabidopsis genome
- PMID: 38609974
- PMCID: PMC11015597
- DOI: 10.1186/s12915-024-01882-5
Nuclear lamina component KAKU4 regulates chromatin states and transcriptional regulation in the Arabidopsis genome
Abstract
Background: The nuclear lamina links the nuclear membrane to chromosomes and plays a crucial role in regulating chromatin states and gene expression. However, current knowledge of nuclear lamina in plants is limited compared to animals and humans.
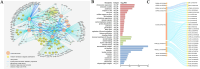
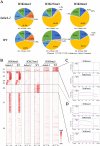
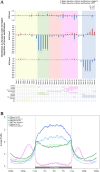
Results: This study mainly focused on elucidating the mechanism through which the putative nuclear lamina component protein KAKU4 regulates chromatin states and gene expression in Arabidopsis leaves. Thus, we constructed a network using the association proteins of lamin-like proteins, revealing that KAKU4 is strongly associated with chromatin or epigenetic modifiers. Then, we conducted ChIP-seq technology to generate global epigenomic profiles of H3K4me3, H3K27me3, and H3K9me2 in Arabidopsis leaves for mutant (kaku4-2) and wild-type (WT) plants alongside RNA-seq method to generate gene expression profiles. The comprehensive chromatin state-based analyses indicate that the knockdown of KAKU4 has the strongest effect on H3K27me3, followed by H3K9me2, and the least impact on H3K4me3, leading to significant changes in chromatin states in the Arabidopsis genome. We discovered that the knockdown of the KAKU4 gene caused a transition between two types of repressive epigenetics marks, H3K9me2 and H3K27me3, in some specific PLAD regions. The combination analyses of epigenomic and transcriptomic data between the kaku4-2 mutant and WT suggested that KAKU4 may regulate key biological processes, such as programmed cell death and hormone signaling pathways, by affecting H3K27me3 modification in Arabidopsis leaves.
Conclusions: In summary, our results indicated that KAKU4 is directly and/or indirectly associated with chromatin/epigenetic modifiers and demonstrated the essential roles of KAKU4 in regulating chromatin states, transcriptional regulation, and diverse biological processes in Arabidopsis.
Keywords: Arabidopsis; Chromatin state; H3K27me3; H3K9me2; Hormone pathway; KAKU4.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
Bath: a Bayesian approach to analyze epigenetic transitions reveals a dual role of H3K27me3 in chondrogenesis.Epigenetics Chromatin. 2025 Jun 27;18(1):38. doi: 10.1186/s13072-025-00594-6. Epigenetics Chromatin. 2025. PMID: 40571950 Free PMC article.
-
KAKU4 regulates leaf senescence through modulation of H3K27me3 deposition in the Arabidopsis genome.BMC Plant Biol. 2024 Mar 7;24(1):177. doi: 10.1186/s12870-024-04860-9. BMC Plant Biol. 2024. PMID: 38448830 Free PMC article.
-
Mutation of the conserved late element in geminivirus CP promoters abolishes Arabidopsis TCP24 transcription factor binding and decreases H3K27me3 levels on viral chromatin.PLoS Pathog. 2024 Jul 18;20(7):e1012399. doi: 10.1371/journal.ppat.1012399. eCollection 2024 Jul. PLoS Pathog. 2024. PMID: 39024402 Free PMC article.
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
References
-
- Betsuyaku S, Katou S, Takebayashi Y, Sakakibara H, Nomura N, Fukuda H. Salicylic acid and jasmonic acid pathways are activated in spatially different domains around the infection site during effector-triggered immunity in Arabidopsis thaliana. Plant Cell Physiol. 2018;59:439. doi: 10.1093/pcp/pcy008. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

