Mutation landscape in Chinese nodal diffuse large B-cell lymphoma by targeted next generation sequencing and their relationship with clinicopathological characteristics
- PMID: 38609996
- PMCID: PMC11015559
- DOI: 10.1186/s12920-024-01866-y
Mutation landscape in Chinese nodal diffuse large B-cell lymphoma by targeted next generation sequencing and their relationship with clinicopathological characteristics
Abstract
Background: Diffuse large B-cell lymphoma (DLBCL), an aggressive and heterogenic malignant entity, is still a challenging clinical problem, since around one-third of patients are not cured with primary treatment. Next-generation sequencing (NGS) technologies have revealed common genetic mutations in DLBCL. We devised an NGS multi-gene panel to discover genetic features of Chinese nodal DLBCL patients and provide reference information for panel-based NGS detection in clinical laboratories.
Methods: A panel of 116 DLBCL genes was designed based on the literature and related databases. We analyzed 96 Chinese nodal DLBCL biopsy specimens through targeted sequencing.
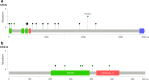
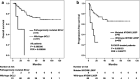
Results: The most frequently mutated genes were KMT2D (30%), PIM1 (26%), SOCS1 (24%), MYD88 (21%), BTG1 (20%), HIST1H1E (18%), CD79B (18%), SPEN (17%), and KMT2C (16%). SPEN (17%) and DDX3X (6%) mutations were highly prevalent in our study than in Western studies. Thirty-three patients (34%) were assigned as genetic classification by the LymphGen algorithm, including 12 cases MCD, five BN2, seven EZB, seven ST2, and two EZB/ST2 complex. MYD88 L265P mutation, TP53 and BCL2 pathogenic mutations were unfavorable prognostic biomarkers in DLBCL.
Conclusions: This study presents the mutation landscape in Chinese nodal DLBCL, highlights the genetic heterogeneity of DLBCL and shows the role of panel-based NGS to prediction of prognosis and potential molecular targeted therapy in DLBCL. More precise genetic classification needs further investigations.
Keywords: Diffuse large B-cell lymphoma; Genetic subtype; Mutation; Next-generation sequencing; Targeted sequencing.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, et al. WHO classification of tumours of haematopoietic and lymphoid tissues, 4th ed. Lyon: International Agency for Research on Cancer (IARC); 2008.
-
- Li X, Li G, Gao Z, Zhou X, Zhu X. The relative frequencies of lymphoma subtypes in China: a nationwide study of 10002 cases by the Chinese lymphoma study group. Ann Oncol. 2011;224:141.
-
- Coiffier B, Thieblemont C, Van Den Neste E, Lepeu G, Plantier I, Castaigne S, et al. Long-term outcome of patients in the LNH-98.5 trial, the first randomized study comparing rituximab-CHOP to standard CHOP chemotherapy in DLBCL patients: a study by the Groupe d'Etudes des Lymphomes de l'Adulte. Blood. 2010;116(12):2040–5. doi: 10.1182/blood-2010-03-276246. - DOI - PMC - PubMed
-
- Pfreundschuh M, Kuhnt E, Trumper L, Osterborg A, Trneny M, Shepherd L, et al. CHOP-like chemotherapy with or without rituximab in young patients with good-prognosis diffuse large-B-cell lymphoma: 6-year results of an open-label randomised study of the MabThera International Trial (MInT) Group. Lancet Oncol. 2011;12(11):1013–1022. doi: 10.1016/S1470-2045(11)70235-2. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

