Transcriptome-Wide N6-Methyladenosine (m6A) Methylation Analyses in a Compatible Wheat- Puccinia striiformis f. sp. tritici Interaction
- PMID: 38611510
- PMCID: PMC11013425
- DOI: 10.3390/plants13070982
Transcriptome-Wide N6-Methyladenosine (m6A) Methylation Analyses in a Compatible Wheat- Puccinia striiformis f. sp. tritici Interaction
Abstract
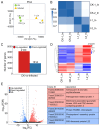
N6-methyladenosine (m6A) is a prevalent internal modification in eukaryotic mRNA, tRNA, miRNA, and long non-coding RNA. It is also known for its role in plant responses to biotic and abiotic stresses. However, a comprehensive m6A transcriptome-wide map for Puccinia striiformis f. sp. tritici (Pst) infections in wheat (Triticum aestivum) is currently unavailable. Our study is the first to profile m6A modifications in wheat infected with a virulent Pst race. Analysis of RNA-seq and MeRIP-seq data revealed that the majority of differentially expressed genes are up-regulated and hyper-methylated. Some of these genes are enriched in the plant-pathogen interaction pathway. Notably, genes related to photosynthesis showed significant down-regulation and hypo-methylation, suggesting a potential mechanism facilitating successful Pst invasion by impairing photosynthetic function. The crucial genes, epitomizing the core molecular constituents that fortify plants against pathogenic assaults, were detected with varying expression and methylation levels, together with a newly identified methylation motif. Additionally, m6A regulator genes were also influenced by m6A modification, and their expression patterns varied at different time points of post-inoculation, with lower expression at early stages of infection. This study provides insights into the role of m6A modification regulation in wheat's response to Pst infection, establishing a foundation for understanding the potential function of m6A RNA methylation in plant resistance or susceptibility to pathogens.
Keywords: MeRIP-seq; Puccinia striiformis f. sp. tritici; RNA-seq; gene expression; m6A RNA methylation; photosynthesis; plant–pathogen interaction; post-transcriptional modification; wheat.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



Similar articles
-
Metabolite profiling of susceptible and resistant wheat (Triticum aestivum) cultivars responding to Puccinia striiformis f. sp. tritici infection.BMC Plant Biol. 2023 Jun 1;23(1):293. doi: 10.1186/s12870-023-04313-9. BMC Plant Biol. 2023. PMID: 37264330 Free PMC article.
-
Puccinia striiformis f. sp. tritici microRNA-like RNA 1 (Pst-milR1), an important pathogenicity factor of Pst, impairs wheat resistance to Pst by suppressing the wheat pathogenesis-related 2 gene.New Phytol. 2017 Jul;215(1):338-350. doi: 10.1111/nph.14577. Epub 2017 May 2. New Phytol. 2017. PMID: 28464281
-
Transcriptome-Wide N6-Methyladenosine (m6A) Profiling of Susceptible and Resistant Wheat Varieties Reveals the Involvement of Variety-Specific m6A Modification Involved in Virus-Host Interaction Pathways.Front Microbiol. 2021 May 26;12:656302. doi: 10.3389/fmicb.2021.656302. eCollection 2021. Front Microbiol. 2021. PMID: 34122371 Free PMC article.
-
Down-regulation of a wheat alkaline/neutral invertase correlates with reduced host susceptibility to wheat stripe rust caused by Puccinia striiformis.J Exp Bot. 2015 Dec;66(22):7325-38. doi: 10.1093/jxb/erv428. Epub 2015 Sep 18. J Exp Bot. 2015. PMID: 26386259
-
Wheat stripe (yellow) rust caused by Puccinia striiformis f. sp. tritici.Mol Plant Pathol. 2014 Jun;15(5):433-46. doi: 10.1111/mpp.12116. Mol Plant Pathol. 2014. PMID: 24373199 Free PMC article. Review.
Cited by
-
RNA modifications in plant biotic interactions.Plant Commun. 2025 Feb 10;6(2):101232. doi: 10.1016/j.xplc.2024.101232. Epub 2024 Dec 25. Plant Commun. 2025. PMID: 39722456 Free PMC article. Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources

