Decrypting Molecular Mechanisms Involved in Counteracting Copper and Nickel Toxicity in Jack Pine (Pinus banksiana) Based on Transcriptomic Analysis
- PMID: 38611570
- PMCID: PMC11013723
- DOI: 10.3390/plants13071042
Decrypting Molecular Mechanisms Involved in Counteracting Copper and Nickel Toxicity in Jack Pine (Pinus banksiana) Based on Transcriptomic Analysis
Abstract
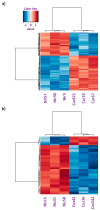
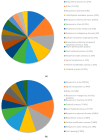
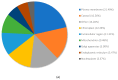
The remediation of copper and nickel-afflicted sites is challenged by the different physiological effects imposed by each metal on a given plant system. Pinus banksiana is resilient against copper and nickel, providing an opportunity to build a valuable resource to investigate the responding gene expression toward each metal. The objectives of this study were to (1) extend the analysis of the Pinus banksiana transcriptome exposed to nickel and copper, (2) assess the differential gene expression in nickel-resistant compared to copper-resistant genotypes, and (3) identify mechanisms specific to each metal. The Illumina platform was used to sequence RNA that was extracted from seedlings treated with each of the metals. There were 449 differentially expressed genes (DEGs) between copper-resistant genotypes (RGs) and nickel-resistant genotypes (RGs) at a high stringency cut-off, indicating a distinct pattern of gene expression toward each metal. For biological processes, 19.8% of DEGs were associated with the DNA metabolic process, followed by the response to stress (13.15%) and the response to chemicals (8.59%). For metabolic function, 27.9% of DEGs were associated with nuclease activity, followed by nucleotide binding (27.64%) and kinase activity (10.16%). Overall, 21.49% of DEGs were localized to the plasma membrane, followed by the cytosol (16.26%) and chloroplast (12.43%). Annotation of the top upregulated genes in copper RG compared to nickel RG identified genes and mechanisms that were specific to copper and not to nickel. NtPDR, AtHIPP10, and YSL1 were identified as genes associated with copper resistance. Various genes related to cell wall metabolism were identified, and they included genes encoding for HCT, CslE6, MPG, and polygalacturonase. Annotation of the top downregulated genes in copper RG compared to nickel RG revealed genes and mechanisms that were specific to nickel and not copper. Various regulatory and signaling-related genes associated with the stress response were identified. They included UGT, TIFY, ACC, dirigent protein, peroxidase, and glyoxyalase I. Additional research is needed to determine the specific functions of signaling and stress response mechanisms in nickel-resistant plants.
Keywords: Pinus banksiana; biological process; cellular compartment; functional genomic; molecular function; nickel and copper toxicity; resistance mechanisms.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




Similar articles
-
Transcriptome Analysis Reveals Changes in Whole Gene Expression, Biological Process, and Molecular Functions Induced by Nickel in Jack Pine (Pinus banksiana).Plants (Basel). 2023 Aug 7;12(15):2889. doi: 10.3390/plants12152889. Plants (Basel). 2023. PMID: 37571042 Free PMC article.
-
Gene expression profiling of Jack Pine (Pinus banksiana) under copper stress: Identification of genes associated with copper resistance.PLoS One. 2024 Mar 7;19(3):e0296027. doi: 10.1371/journal.pone.0296027. eCollection 2024. PLoS One. 2024. PMID: 38452110 Free PMC article.
-
Transcriptome analysis of trembling aspen (Populus tremuloides) under nickel stress.PLoS One. 2022 Oct 13;17(10):e0274740. doi: 10.1371/journal.pone.0274740. eCollection 2022. PLoS One. 2022. PMID: 36227867 Free PMC article.
-
Transcriptome analysis of Pinus monticola primary needles by RNA-seq provides novel insight into host resistance to Cronartium ribicola.BMC Genomics. 2013 Dec 16;14:884. doi: 10.1186/1471-2164-14-884. BMC Genomics. 2013. PMID: 24341615 Free PMC article.
-
Differential levels of gene expression and molecular mechanisms between red maple (Acer rubrum) genotypes resistant and susceptible to nickel toxicity revealed by transcriptome analysis.Ecol Evol. 2018 Apr 19;8(10):4876-4890. doi: 10.1002/ece3.4045. eCollection 2018 May. Ecol Evol. 2018. PMID: 29876066 Free PMC article.
References
-
- DeForest D.K., Brix K.V., Adams W.J. Assessing Metal Bioaccumulation in Aquatic Environments: The Inverse Relationship between Bioaccumulation Factors, Trophic Transfer Factors and Exposure Concentration. Aquat. Toxicol. Amst. Neth. 2007;84:236–246. doi: 10.1016/j.aquatox.2007.02.022. - DOI - PubMed
-
- Jewiss T. The Mining History of the Sudbury Area|Earth Sciences Museum. [(accessed on 1 January 2022)]. Available online: https://uwaterloo.ca/earth-sciences-museum/resources/mining-canada/minin....
LinkOut - more resources
Full Text Sources

