Regulating Protein-RNA Interactions: Advances in Targeting the LIN28/Let-7 Pathway
- PMID: 38612395
- PMCID: PMC11011352
- DOI: 10.3390/ijms25073585
Regulating Protein-RNA Interactions: Advances in Targeting the LIN28/Let-7 Pathway
Abstract
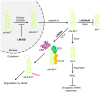
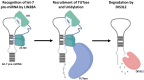
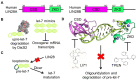
Originally discovered in C. elegans, LIN28 is an evolutionarily conserved zinc finger RNA-binding protein (RBP) that post-transcriptionally regulates genes involved in developmental timing, stem cell programming, and oncogenesis. LIN28 acts via two distinct mechanisms. It blocks the biogenesis of the lethal-7 (let-7) microRNA (miRNA) family, and also directly binds messenger RNA (mRNA) targets, such as IGF-2 mRNA, and alters downstream splicing and translation events. This review focuses on the molecular mechanism of LIN28 repression of let-7 and current strategies to overcome this blockade for the purpose of cancer therapy. We highlight the value of the LIN28/let-7 pathway as a drug target, as multiple oncogenic proteins that the pathway regulates are considered undruggable due to their inaccessible cellular location and lack of cavities for small molecule binding.
Keywords: LIN28; gene therapy; let-7; miRNA; oncogene; small-molecule inhibitor.
Conflict of interest statement
P.S. holds positions and intellectual property at 28/7 Therapeutics, a company aimed at developing inhibitors of the LIN28/let-7 pathway.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

