Properties and Mechanisms of Deletions, Insertions, and Substitutions in the Evolutionary History of SARS-CoV-2
- PMID: 38612505
- PMCID: PMC11011937
- DOI: 10.3390/ijms25073696
Properties and Mechanisms of Deletions, Insertions, and Substitutions in the Evolutionary History of SARS-CoV-2
Abstract
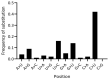
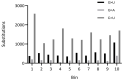
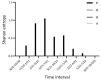
SARS-CoV-2 has accumulated many mutations since its emergence in late 2019. Nucleotide substitutions leading to amino acid replacements constitute the primary material for natural selection. Insertions, deletions, and substitutions appear to be critical for coronavirus's macro- and microevolution. Understanding the molecular mechanisms of mutations in the mutational hotspots (positions, loci with recurrent mutations, and nucleotide context) is important for disentangling roles of mutagenesis and selection. In the SARS-CoV-2 genome, deletions and insertions are frequently associated with repetitive sequences, whereas C>U substitutions are often surrounded by nucleotides resembling the APOBEC mutable motifs. We describe various approaches to mutation spectra analyses, including the context features of RNAs that are likely to be involved in the generation of recurrent mutations. We also discuss the interplay between mutations and natural selection as a complex evolutionary trend. The substantial variability and complexity of pipelines for the reconstruction of mutations and the huge number of genomic sequences are major problems for the analyses of mutations in the SARS-CoV-2 genome. As a solution, we advocate for the development of a centralized database of predicted mutations, which needs to be updated on a regular basis.
Keywords: ADAR; APOBEC; SARS-CoV-2; epistasis; low-complexity regions; mutation hotspots; oxidative stress; viral fitness.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

