An Overview of the Epigenetic Modifications in the Brain under Normal and Pathological Conditions
- PMID: 38612690
- PMCID: PMC11011998
- DOI: 10.3390/ijms25073881
An Overview of the Epigenetic Modifications in the Brain under Normal and Pathological Conditions
Abstract
Epigenetic changes are changes in gene expression that do not involve alterations to the DNA sequence. These changes lead to establishing a so-called epigenetic code that dictates which and when genes are activated, thus orchestrating gene regulation and playing a central role in development, health, and disease. The brain, being mostly formed by cells that do not undergo a renewal process throughout life, is highly prone to the risk of alterations leading to neuronal death and neurodegenerative disorders, mainly at a late age. Here, we review the main epigenetic modifications that have been described in the brain, with particular attention on those related to the onset of developmental anomalies or neurodegenerative conditions and/or occurring in old age. DNA methylation and several types of histone modifications (acetylation, methylation, phosphorylation, ubiquitination, sumoylation, lactylation, and crotonylation) are major players in these processes. They are directly or indirectly involved in the onset of neurodegeneration in Alzheimer's or Parkinson's disease. Therefore, this review briefly describes the roles of these epigenetic changes in the mechanisms of brain development, maturation, and aging and some of the most important factors dynamically regulating or contributing to these changes, such as oxidative stress, inflammation, and mitochondrial dysfunction.
Keywords: DNA; brain; development; epigenetics; histones; neurodegeneration; neurons.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
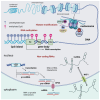
References
-
- Pérez R.F., Santamarina P., Fernández A.F., Fraga M.F. Chapter 19—Epigenetics and Lifestyle: The Impact of Stress, Diet, and Social Habits on Tissue Homeostasis. In: Palacios D., editor. Epigenetics and Regeneration. Volume 11. Academic Press; Cambridge, MA, USA: 2019. pp. 461–489.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

