Claudin-10 Expression and the Gene Expression Pattern of Thick Ascending Limb Cells
- PMID: 38612818
- PMCID: PMC11011785
- DOI: 10.3390/ijms25074008
Claudin-10 Expression and the Gene Expression Pattern of Thick Ascending Limb Cells
Abstract
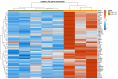
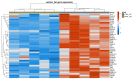
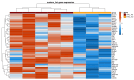
Many genomic, anatomical and functional differences exist between the medullary (MTAL) and the cortical thick ascending limb of the loop of Henle (CTAL), including a higher expression of claudin-10 (CLDN10) in the MTAL than in the CTAL. Therefore, we assessed to what extent the Cldn10 gene expression is a determinant of differential gene expression between MTAL and CTAL. RNAs extracted from CTAL and MTAL microdissected from wild type (WT) and Cldn10 knock out mice (cKO) were analyzed by RNAseq. Differential and enrichment analyses (GSEA) were performed with interactive R Shiny software. Between WT and cKO MTAL, 637 genes were differentially expressed, whereas only 76 were differentially expressed between WT and cKO CTAL. Gene expression patterns and GSEA analyses in all replicates showed that WT MTAL did not cluster with the other replicates; no hierarchical clustering could be found between WT CTAL, cKO CTAL and cKO MTAL. Compared to WT replicates, cKO replicates were enriched in Cldn16, Cldn19, Pth1r, (parathyroid hormone receptor type 1), Casr (calcium sensing receptor) and Vdr (Vitamin D Receptor) mRNA in both the cortex and medulla. Cldn10 is associated with gene expression patterns, including genes specifically involved in divalent cations reabsorption in the TAL.
Keywords: HELIX syndrome; claudin-10; epithelium; kidney; thick ascending limb of the loop of Henle; tight junction; transcriptional profiling.
Conflict of interest statement
P.H. and C.P.-B. have no conflicts of interest to declare regarding the content of the present study.
Figures






References
-
- Konrad M., Nijenhuis T., Ariceta G., Bertholet-Thomas A., Calo L.A., Capasso G., Emma F., Schlingmann K.P., Singh M., Trepiccione F., et al. Diagnosis and management of Bartter syndrome: Executive summary of the consensus and recommendations from the European Rare Kidney Disease Reference Network Working Group for Tubular Disorders. Kidney Int. 2021;99:324–335. doi: 10.1016/j.kint.2020.10.035. - DOI - PubMed
-
- Bongers E., Shelton L.M., Milatz S., Verkaart S., Bech A.P., Schoots J., Cornelissen E.A.M., Bleich M., Hoenderop J.G.J., Wetzels J.F.M., et al. A Novel Hypokalemic-Alkalotic Salt-Losing Tubulopathy in Patients with CLDN10 Mutations. J. Am. Soc. Nephrol. 2017;28:3118–3128. doi: 10.1681/ASN.2016080881. - DOI - PMC - PubMed
-
- Klar J., Piontek J., Milatz S., Tariq M., Jameel M., Breiderhoff T., Schuster J., Fatima A., Asif M., Sher M., et al. Altered paracellular cation permeability due to a rare CLDN10B variant causes anhidrosis and kidney damage. PLoS Genet. 2017;13:e1006897. doi: 10.1371/journal.pgen.1006897. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

