Transcriptome Analysis of Tomato Leaves Reveals Candidate Genes Responsive to Tomato Brown Rugose Fruit Virus Infection
- PMID: 38612822
- PMCID: PMC11012278
- DOI: 10.3390/ijms25074012
Transcriptome Analysis of Tomato Leaves Reveals Candidate Genes Responsive to Tomato Brown Rugose Fruit Virus Infection
Abstract
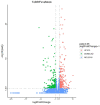
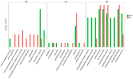
Tomato brown rugose fruit virus (ToBRFV) is a newly-emerging tobamovirus which was first reported on tomatoes in Israel and Jordan, and which has now spread rapidly in Asia, Europe, North America, and Africa. ToBRFV can overcome the resistance to other tobamoviruses conferred by tomato Tm-1, Tm-2, and Tm-22 genes, and it has seriously affected global crop production. The rapid and comprehensive transcription reprogramming of host plant cells is the key to resisting virus attack, but there have been no studies of the transcriptome changes induced by ToBRFV in tomatoes. Here, we made a comparative transcriptome analysis between tomato leaves infected with ToBRFV for 21 days and those mock-inoculated as controls. A total of 522 differentially expressed genes were identified after ToBRFV infection, of which 270 were up-regulated and 252 were down-regulated. Functional analysis showed that DEGs were involved in biological processes such as response to wounding, response to stress, protein folding, and defense response. Ten DEGs were selected and verified by qRT-PCR, confirming the reliability of the high-throughput sequencing data. These results provide candidate genes or signal pathways for the response of tomato leaves to ToBRFV infection.
Keywords: tomato; tomato brown rugose fruit virus; transcriptome analysis.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures



Similar articles
-
Tomato brown rugose fruit virus: An emerging and rapidly spreading plant RNA virus that threatens tomato production worldwide.Mol Plant Pathol. 2022 Sep;23(9):1262-1277. doi: 10.1111/mpp.13229. Epub 2022 May 22. Mol Plant Pathol. 2022. PMID: 35598295 Free PMC article. Review.
-
Comparative Analysis of Host Range, Ability to Infect Tomato Cultivars with Tm-22 Gene, and Real-Time Reverse Transcription PCR Detection of Tomato Brown Rugose Fruit Virus.Plant Dis. 2021 Nov;105(11):3643-3652. doi: 10.1094/PDIS-05-20-1070-RE. Epub 2021 Nov 18. Plant Dis. 2021. PMID: 34058839
-
The Tomato Brown Rugose Fruit Virus Movement Protein Overcomes Tm-22 Resistance in Tomato While Attenuating Viral Transport.Mol Plant Microbe Interact. 2021 Sep;34(9):1024-1032. doi: 10.1094/MPMI-01-21-0023-R. Epub 2021 Oct 11. Mol Plant Microbe Interact. 2021. PMID: 33970669
-
Identification of genetic determinants of tomato brown rugose fruit virus that enable infection of plants harbouring the Tm-22 resistance gene.Mol Plant Pathol. 2021 Nov;22(11):1347-1357. doi: 10.1111/mpp.13115. Epub 2021 Aug 13. Mol Plant Pathol. 2021. PMID: 34390124 Free PMC article.
-
Tomato Brown Rugose Fruit Virus Pandemic.Annu Rev Phytopathol. 2023 Sep 5;61:137-164. doi: 10.1146/annurev-phyto-021622-120703. Epub 2023 Jun 2. Annu Rev Phytopathol. 2023. PMID: 37268006 Review.
Cited by
-
Unveiling the Role of SlRNC1 in Chloroplast Development and Global Gene Regulation in Tomato Plants.Int J Mol Sci. 2024 Jun 24;25(13):6898. doi: 10.3390/ijms25136898. Int J Mol Sci. 2024. PMID: 39000008 Free PMC article.
-
Transcriptome Analysis of Pepper Leaves in Response to Tomato Brown Rugose Fruit Virus Infection.Plants (Basel). 2025 Apr 23;14(9):1280. doi: 10.3390/plants14091280. Plants (Basel). 2025. PMID: 40364309 Free PMC article.
-
Omics based approaches to decipher the leaf ionome and transcriptome changes in Solanum lycopersicum L. upon Tomato Brown Rugose Fruit Virus (ToBRFV) infection.PLoS One. 2024 Nov 8;19(11):e0313335. doi: 10.1371/journal.pone.0313335. eCollection 2024. PLoS One. 2024. PMID: 39514546 Free PMC article.
References
-
- Hanssen I.M., Lapidot M. Major tomato viruses in the Mediterranean basin. Adv. Virus Res. 2012;84:31–66. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

