Biosynthesis of Hesperetin, Homoeriodictyol, and Homohesperetin in a Transcriptomics-Driven Engineered Strain of Streptomyces albidoflavus
- PMID: 38612864
- PMCID: PMC11012174
- DOI: 10.3390/ijms25074053
Biosynthesis of Hesperetin, Homoeriodictyol, and Homohesperetin in a Transcriptomics-Driven Engineered Strain of Streptomyces albidoflavus
Abstract
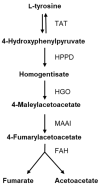
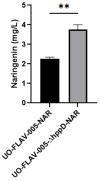
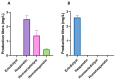
Flavonoids exhibit various bioactivities including anti-oxidant, anti-tumor, anti-inflammatory, and anti-viral properties. Methylated flavonoids are particularly significant due to their enhanced oral bioavailability, improved intestinal absorption, and greater stability. The heterologous production of plant flavonoids in bacterial factories involves the need for enough biosynthetic precursors to allow for high production levels. These biosynthetic precursors are malonyl-CoA and l-tyrosine. In this work, to enhance flavonoid biosynthesis in Streptomyces albidoflavus, we conducted a transcriptomics study for the identification of candidate genes involved in l-tyrosine catabolism. The hypothesis was that the bacterial metabolic machinery would detect an excess of this amino acid if supplemented with the conventional culture medium and would activate the genes involved in its catabolism towards energy production. Then, by inactivating those overexpressed genes (under an excess of l-tyrosine), it would be possible to increase the intracellular pools of this precursor amino acid and eventually the final flavonoid titers in this bacterial factory. The RNAseq data analysis in the S. albidoflavus wild-type strain highlighted the hppD gene encoding 4-hydroxyphenylpyruvate dioxygenase as a promising target for knock-out, exhibiting a 23.2-fold change (FC) in expression upon l-tyrosine supplementation in comparison to control cultivation conditions. The subsequent knock-out of the hppD gene in S. albidoflavus resulted in a 1.66-fold increase in the naringenin titer, indicating enhanced flavonoid biosynthesis. Leveraging the improved strain of S. albidoflavus, we successfully synthesized the methylated flavanones hesperetin, homoeriodictyol, and homohesperetin, achieving titers of 2.52 mg/L, 1.34 mg/L, and 0.43 mg/L, respectively. In addition, the dimethoxy flavanone homohesperetin was produced as a byproduct of the endogenous metabolism of S. albidoflavus. To our knowledge, this is the first time that hppD deletion was utilized as a strategy to augment the biosynthesis of flavonoids. Furthermore, this is the first report where hesperetin and homoeriodictyol have been synthesized from l-tyrosine as a precursor. Therefore, transcriptomics is, in this case, a successful approach for the identification of catabolism reactions affecting key precursors during flavonoid biosynthesis, allowing the generation of enhanced production strains.
Keywords: flavonoid; l-tyrosine feeding; methyltransferase; substrate flexibility.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




Similar articles
-
Identification of a polyphenol O-methyltransferase with broad substrate flexibility in Streptomyces albidoflavus J1074.Microb Cell Fact. 2024 Oct 5;23(1):265. doi: 10.1186/s12934-024-02541-8. Microb Cell Fact. 2024. PMID: 39369216 Free PMC article.
-
Metabolic engineering in Streptomyces albidoflavus for the biosynthesis of the methylated flavonoids sakuranetin, acacetin, and genkwanin.Microb Cell Fact. 2023 Nov 14;22(1):234. doi: 10.1186/s12934-023-02247-3. Microb Cell Fact. 2023. PMID: 37964284 Free PMC article.
-
Use of 3-Deoxy-D-arabino-heptulosonic acid 7-phosphate Synthase (DAHP Synthase) to Enhance the Heterologous Biosynthesis of Diosmetin and Chrysoeriol in an Engineered Strain of Streptomyces albidoflavus.Int J Mol Sci. 2024 Feb 28;25(5):2776. doi: 10.3390/ijms25052776. Int J Mol Sci. 2024. PMID: 38474023 Free PMC article.
-
Systems metabolic engineering of the primary and secondary metabolism of Streptomyces albidoflavus enhances production of the reverse antibiotic nybomycin against multi-resistant Staphylococcus aureus.Metab Eng. 2024 Jan;81:123-143. doi: 10.1016/j.ymben.2023.12.004. Epub 2023 Dec 9. Metab Eng. 2024. PMID: 38072358
-
Heterologous production of flavanones in Escherichia coli: potential for combinatorial biosynthesis of flavonoids in bacteria.J Ind Microbiol Biotechnol. 2003 Aug;30(8):456-61. doi: 10.1007/s10295-003-0061-1. Epub 2003 May 21. J Ind Microbiol Biotechnol. 2003. PMID: 12759810 Review.
Cited by
-
Identification of a polyphenol O-methyltransferase with broad substrate flexibility in Streptomyces albidoflavus J1074.Microb Cell Fact. 2024 Oct 5;23(1):265. doi: 10.1186/s12934-024-02541-8. Microb Cell Fact. 2024. PMID: 39369216 Free PMC article.
References
-
- Rainey-Smith S., Schroetke L.-W., Bahia P., Fahmi A., Skilton R., Spencer J.P.E., Rice-Evans C., Rattray M., Williams R.J. Neuroprotective Effects of Hesperetin in Mouse Primary Neurones Are Independent of CREB Activation. Neurosci. Lett. 2008;438:29–33. doi: 10.1016/j.neulet.2008.04.056. - DOI - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

