Characterization of the First Secreted Sorting Nexin Identified in the Leishmania Protists
- PMID: 38612903
- PMCID: PMC11012638
- DOI: 10.3390/ijms25074095
Characterization of the First Secreted Sorting Nexin Identified in the Leishmania Protists
Abstract
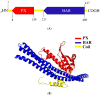
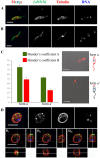
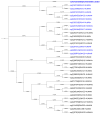
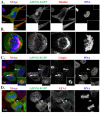
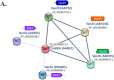
Proteins of the sorting nexin (SNX) family present a modular structural architecture with a phox homology (PX) phosphoinositide (PI)-binding domain and additional PX structural domains, conferring to them a wide variety of vital eukaryotic cell's functions, from signal transduction to membrane deformation and cargo binding. Although SNXs are well studied in human and yeasts, they are poorly investigated in protists. Herein, is presented the characterization of the first SNX identified in Leishmania protozoan parasites encoded by the LdBPK_352470 gene. In silico secondary and tertiary structure prediction revealed a PX domain on the N-terminal half and a Bin/amphiphysin/Rvs (BAR) domain on the C-terminal half of this protein, with these features classifying it in the SNX-BAR subfamily of SNXs. We named the LdBPK_352470.1 gene product LdSNXi, as it is the first SNX identified in Leishmania (L.) donovani. Its expression was confirmed in L. donovani promastigotes under different cell cycle phases, and it was shown to be secreted in the extracellular medium. Using an in vitro lipid binding assay, it was demonstrated that recombinant (r) LdSNXi (rGST-LdSNXi) tagged with glutathione-S-transferase (GST) binds to the PtdIns3P and PtdIns4P PIs. Using a specific a-LdSNXi antibody and immunofluorescence confocal microscopy, the intracellular localization of endogenous LdSNXi was analyzed in L. donovani promastigotes and axenic amastigotes. Additionally, rLdSNXi tagged with enhanced green fluorescent protein (rLdSNXi-EGFP) was heterologously expressed in transfected HeLa cells and its localization was examined. All observed localizations suggest functions compatible with the postulated SNX identity of LdSNXi. Sequence, structure, and evolutionary analysis revealed high homology between LdSNXi and the human SNX2, while the investigation of protein-protein interactions based on STRING (v.11.5) predicted putative molecular partners of LdSNXi in Leishmania.
Keywords: BAR domain; Leishmania donovani; PX domain; SNX-BAR; SNX2; phosphoinositide binding protein; sorting nexin.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
A large family of endosome-localized proteins related to sorting nexin 1.Biochem J. 2001 Aug 15;358(Pt 1):7-16. doi: 10.1042/0264-6021:3580007. Biochem J. 2001. PMID: 11485546 Free PMC article.
-
SNX-PXA-RGS-PXC Subfamily of SNXs in the Regulation of Receptor-Mediated Signaling and Membrane Trafficking.Int J Mol Sci. 2021 Feb 26;22(5):2319. doi: 10.3390/ijms22052319. Int J Mol Sci. 2021. PMID: 33652569 Free PMC article. Review.
-
Structural basis for different phosphoinositide specificities of the PX domains of sorting nexins regulating G-protein signaling.J Biol Chem. 2014 Oct 10;289(41):28554-68. doi: 10.1074/jbc.M114.595959. Epub 2014 Aug 22. J Biol Chem. 2014. PMID: 25148684 Free PMC article.
-
Structure and Membrane Binding Properties of the Endosomal Tetratricopeptide Repeat (TPR) Domain-containing Sorting Nexins SNX20 and SNX21.J Biol Chem. 2015 Jun 5;290(23):14504-17. doi: 10.1074/jbc.M115.650598. Epub 2015 Apr 16. J Biol Chem. 2015. PMID: 25882846 Free PMC article.
-
The Phox Homology (PX) Domain.Adv Exp Med Biol. 2019;1111:1-17. doi: 10.1007/5584_2018_185. Adv Exp Med Biol. 2019. PMID: 29569114 Review.
Cited by
-
The role of AMPK in pancreatic cancer: from carcinogenesis to treatment.Clin Transl Oncol. 2025 Jan;27(1):70-82. doi: 10.1007/s12094-024-03572-8. Epub 2024 Jun 26. Clin Transl Oncol. 2025. PMID: 38926257 Review.
References
-
- Chishti A.H., Kim A.C., Marfatia S.M., Lutchman M., Hanspal M., Jindal H., Liu S.-C., Low P.S., Rouleau G.A., Mohandas N., et al. The FERM domain: A unique module involved in the linkage of cytoplasmic proteins to the membrane. Trends Biochem. Sci. 1998;23:281–282. doi: 10.1016/S0968-0004(98)01237-7. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

