NeoAgDT: optimization of personal neoantigen vaccine composition by digital twin simulation of a cancer cell population
- PMID: 38614133
- PMCID: PMC11076149
- DOI: 10.1093/bioinformatics/btae205
NeoAgDT: optimization of personal neoantigen vaccine composition by digital twin simulation of a cancer cell population
Abstract
Motivation: Neoantigen vaccines make use of tumor-specific mutations to enable the patient's immune system to recognize and eliminate cancer. Selecting vaccine elements, however, is a complex task which needs to take into account not only the underlying antigen presentation pathway but also tumor heterogeneity.
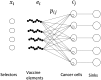
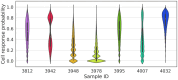
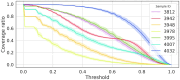
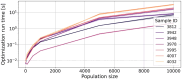
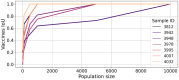
Results: Here, we present NeoAgDT, a two-step approach consisting of: (i) simulating individual cancer cells to create a digital twin of the patient's tumor cell population and (ii) optimizing the vaccine composition by integer linear programming based on this digital twin. NeoAgDT shows improved selection of experimentally validated neoantigens over ranking-based approaches in a study of seven patients.
Availability and implementation: The NeoAgDT code is published on Github: https://github.com/nec-research/neoagdt.
© The Author(s) 2024. Published by Oxford University Press.
Conflict of interest statement
A.M. and P.M. are employees of NEC Laboratories GmbH. F.G. and B.M. were employees of NEC Laboratories GmbH at the time the research was conducted.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

