Spatial and single-cell colocalisation analysis reveals MDK-mediated immunosuppressive environment with regulatory T cells in colorectal carcinogenesis
- PMID: 38614865
- PMCID: PMC11121171
- DOI: 10.1016/j.ebiom.2024.105102
Spatial and single-cell colocalisation analysis reveals MDK-mediated immunosuppressive environment with regulatory T cells in colorectal carcinogenesis
Abstract
Background: Cell-cell interaction factors that facilitate the progression of adenoma to sporadic colorectal cancer (CRC) remain unclear, thereby hindering patient survival.
Methods: We performed spatial transcriptomics on five early CRC cases, which included adenoma and carcinoma, and one advanced CRC. To elucidate cell-cell interactions within the tumour microenvironment (TME), we investigated the colocalisation network at single-cell resolution using a deep generative model for colocalisation analysis, combined with a single-cell transcriptome, and assessed the clinical significance in CRC patients.
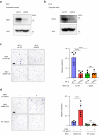
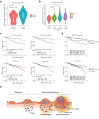
Findings: CRC cells colocalised with regulatory T cells (Tregs) at the adenoma-carcinoma interface. At early-stage carcinogenesis, cell-cell interaction inference between colocalised adenoma and cancer epithelial cells and Tregs based on the spatial distribution of single cells highlighted midkine (MDK) as a prominent signalling molecule sent from tumour epithelial cells to Tregs. Interaction between MDK-high CRC cells and SPP1+ macrophages and stromal cells proved to be the mechanism underlying immunosuppression in the TME. Additionally, we identified syndecan4 (SDC4) as a receptor for MDK associated with Treg colocalisation. Finally, clinical analysis using CRC datasets indicated that increased MDK/SDC4 levels correlated with poor overall survival in CRC patients.
Interpretation: MDK is involved in the immune tolerance shown by Tregs to tumour growth. MDK-mediated formation of the TME could be a potential target for early diagnosis and treatment of CRC.
Funding: Japan Society for the Promotion of Science (JSPS) Grant-in-Aid for Science Research; OITA Cancer Research Foundation; AMED under Grant Number; Japan Science and Technology Agency (JST); Takeda Science Foundation; The Princess Takamatsu Cancer Research Fund.
Keywords: Colorectal cancer; Immune tolerance; MDK; SDC4; Single-cell RNA sequencing; Spatial transcriptomics.
Copyright © 2024. Published by Elsevier B.V.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures





References
-
- Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. - PubMed
-
- Lichtenstein P., Holm N.V., Verkasalo P.K., et al. Environmental and heritable factors in the causation of cancer--analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med. 2000;343(2):78–85. - PubMed
-
- Leslie A., Carey F.A., Pratt N.R., Steele R.J. The colorectal adenoma-carcinoma sequence. Br J Surg. 2002;89(7):845–860. - PubMed
-
- Bond J.H. Clinical evidence for the adenoma-carcinoma sequence, and the management of patients with colorectal adenomas. Semin Gastrointest Dis. 2000;11:176–184. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

