NKp44/HLA-DP-dependent regulation of CD8 effector T cells by NK cells
- PMID: 38615318
- PMCID: PMC11416720
- DOI: 10.1016/j.celrep.2024.114089
NKp44/HLA-DP-dependent regulation of CD8 effector T cells by NK cells
Abstract
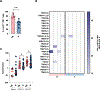
Although natural killer (NK) cells are recognized for their modulation of immune responses, the mechanisms by which human NK cells mediate immune regulation are unclear. Here, we report that expression of human leukocyte antigen (HLA)-DP, a ligand for the activating NK cell receptor NKp44, is significantly upregulated on CD8+ effector T cells, in particular in human cytomegalovirus (HCMV)+ individuals. HLA-DP+ CD8+ T cells expressing NKp44-binding HLA-DP antigens activate NKp44+ NK cells, while HLA-DP+ CD8+ T cells not expressing NKp44-binding HLA-DP antigens do not. In line with this, frequencies of HLA-DP+ CD8+ T cells are increased in individuals not encoding for NKp44-binding HLA-DP haplotypes, and contain hyper-expanded CD8+ T cell clones, compared to individuals expressing NKp44-binding HLA-DP molecules. These findings identify a molecular interaction facilitating the HLA-DP haplotype-specific editing of HLA-DP+ CD8+ T cell effector populations by NKp44+ NK cells and preventing the generation of hyper-expanded T cell clones, which have been suggested to have increased potential for autoimmunity.
Keywords: CD8 cells; CP: Immunology; HCMV; HLA-DP; NK cells; NKp44.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

