This is a preprint.
The structure of the monobactam-producing thioesterase domain of SulM forms a unique complex with the upstream carrier protein domain
- PMID: 38617275
- PMCID: PMC11014566
- DOI: 10.1101/2024.04.06.588331
The structure of the monobactam-producing thioesterase domain of SulM forms a unique complex with the upstream carrier protein domain
Update in
-
The structure of the monobactam-producing thioesterase domain of SulM forms a unique complex with the upstream carrier protein domain.J Biol Chem. 2024 Aug;300(8):107489. doi: 10.1016/j.jbc.2024.107489. Epub 2024 Jun 20. J Biol Chem. 2024. PMID: 38908753 Free PMC article.
Abstract
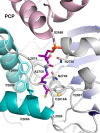
Nonribosomal peptide synthetases (NRPSs) are responsible for the production of important biologically active peptides. The large, multidomain NRPSs operate through an assembly line strategy in which the growing peptide is tethered to carrier domains that deliver the intermediates to neighboring catalytic domains. While most NRPS domains catalyze standard chemistry of amino acid activation, peptide bond formation and product release, some canonical NRPS catalytic domains promote unexpected chemistry. The paradigm monobactam antibiotic sulfazecin is produced through the activity of a terminal thioesterase domain that catalyzes an unusual β-lactam forming reaction in which the nitrogen of the C-terminal N-sulfo-2,3-diaminopropionate residue attacks its thioester tether to release the β-lactam product. We have determined the structure of the thioesterase domain as both a free-standing domain and a didomain complex with the upstream holo peptidyl-carrier domain. The structure illustrates a constrained active site that orients the substrate properly for β-lactam formation. In this regard, the structure is similar to the β-lactone forming thioesterase domain responsible for the production of obafluorin. Analysis of the structure identifies features that are responsible for this four-membered ring closure and enable bioinformatic analysis to identify additional, uncharacterized β-lactam-forming biosynthetic gene clusters by genome mining.
Keywords: NRPS; Nonribosomal Peptide Synthetase; thioesterase; β-lactam antibiotic.
Conflict of interest statement
Conflict of Interest Statement. The authors declare no conflicts of interest with the contents of this article.
Figures






References
-
- Terlouw B. R., Blin K., Navarro-Munoz J. C., Avalon N. E., Chevrette M. G., Egbert S., Lee S., Meijer D., Recchia M. J. J., Reitz Z. L., van Santen J. A., Selem-Mojica N., Torring T., Zaroubi L., Alanjary M., Aleti G., Aguilar C., Al-Salihi S. A. A., Augustijn H. E., Avelar-Rivas J. A., Avitia-Dominguez L. A., Barona-Gomez F., Bernaldo-Aguero J., Bielinski V. A., Biermann F., Booth T. J., Bravo V. J. C., Castelo-Branco R., Chagas F. O., Cruz-Morales P., Gavriilidou A., Gayrard D., Gutierrez-Garcia K., Haslinger K., Helfrich E. J. N., van der Hooft J. J. J., Jati A. P., Kalkreuter E., Kalyvas N., Kang K. B., Kautsar S., Kim W., Kunjapur A. M., Li Y. X., Lin G. M., Loureiro C., Louwen J. J. R., Louwen N. L. L., Lund G., Parra J., Philmus B., Pourmohsenin B., Pronk L. J. U., Rego A., Rex D. A. B., Robinson S., Rosas-Becerra L. R., Roxborough E. T., Schorn M. A., Scobie D. J., Singh K. S., Sokolova N., Tang X. Y., Udwary D., Vigneshwari A., Vind K., Vromans S. P. J. M., Waschulin V., Williams S. E., Winter J. M., Witte T. E., Xie H. L., Yang D., Yu J. W., Zdouc M., Zhong Z., Collemare J., Linington R. G., Weber T., and Medema M. H. (2022) MIBiG 3.0: a community-driven effort to annotate experimentally validated biosynthetic gene clusters. Nucleic Acids Research 51, D603–D610 - PMC - PubMed
-
- Süssmuth R. D., and Mainz A. (2017) Nonribosomal Peptide Synthesis-Principles and Prospects. Angew Chem Int Ed Engl 56, 3770–3821 - PubMed
-
- Medema M. H. (2021) The year 2020 in natural product bioinformatics: an overview of the latest tools and databases. Nat Prod Rep 38, 301–306 - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
