PTPLAD1 Regulates PHB-Raf Interaction to Orchestrate Epithelial-Mesenchymal and Mitofusion-Fission Transitions in Colorectal Cancer
- PMID: 38617530
- PMCID: PMC11008263
- DOI: 10.7150/ijbs.82361
PTPLAD1 Regulates PHB-Raf Interaction to Orchestrate Epithelial-Mesenchymal and Mitofusion-Fission Transitions in Colorectal Cancer
Abstract
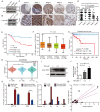
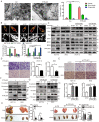
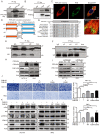
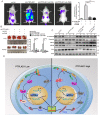
Colorectal cancer (CRC) remains one of the leading causes of cancer-related death worldwide. The poor prognosis of this malignancy is attributed mainly to the persistent activation of cancer signaling for metastasis. Here, we showed that protein tyrosine phosphatase-like A domain containing 1 (PTPLAD1) is down-regulated in highly metastatic CRC cells and negatively associated with poor survival of CRC patients. Systematic analysis reveals that epithelial-to-mesenchymal transition (EMT) and mitochondrial fusion-to-fission (MFT) transition are two critical features for CRC patients with low expression of PTPLAD1. PTPLAD1 overexpression suppresses the metastasis of CRC in vivo and in vitro by inhibiting the Raf/ERK signaling-mediated EMT and mitofission. Mechanically, PTPLAD1 binds with PHB via its middle fragment (141-178 amino acids) and induces dephosphorylation of PHB-Y259 to disrupt the interaction of PHB-Raf, resulting in the inactivation of Raf/ERK signaling. Our results unveil a novel mechanism in which Raf/ERK signaling activated in metastatic CRC induces EMT and mitochondrial fission simultaneously, which can be suppressed by PTPLAD1. This finding may provide a new paradigm for developing more effective treatment strategies for CRC.
Keywords: Colorectal cancer; ERK; Epithelial-to-mesenchymal transition; Metastasis; Mitofission; PHB; PTPLAD1; Raf.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures



References
-
- Courtois G, Gilmore TD. Mutations in the NF-kappaB signaling pathway: implications for human disease. Oncogene. 2006;25:6831–43. - PubMed
-
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A. et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209–49. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

