Early recovery of proteasome activity in cells pulse-treated with proteasome inhibitors is independent of DDI2
- PMID: 38619391
- PMCID: PMC11018354
- DOI: 10.7554/eLife.91678
Early recovery of proteasome activity in cells pulse-treated with proteasome inhibitors is independent of DDI2
Abstract
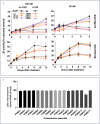
Rapid recovery of proteasome activity may contribute to intrinsic and acquired resistance to FDA-approved proteasome inhibitors. Previous studies have demonstrated that the expression of proteasome genes in cells treated with sub-lethal concentrations of proteasome inhibitors is upregulated by the transcription factor Nrf1 (NFE2L1), which is activated by a DDI2 protease. Here, we demonstrate that the recovery of proteasome activity is DDI2-independent and occurs before transcription of proteasomal genes is upregulated but requires protein translation. Thus, mammalian cells possess an additional DDI2 and transcription-independent pathway for the rapid recovery of proteasome activity after proteasome inhibition.
Keywords: Nrf1; aspartic protease; cancer biology; cell biology; human; ubiquitin; ubiquitin-binding protein; ubl doman.
© 2023, Ibtisam and Kisselev.
Conflict of interest statement
II No competing interests declared, AK AFK is a founder and Chief Scientific Officer of InhiProt LLC
Figures




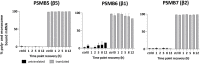

Update of
-
Early recovery of proteasome activity in cells pulse-treated with proteasome inhibitors is independent of DDI2.bioRxiv [Preprint]. 2023 Nov 28:2023.08.03.550647. doi: 10.1101/2023.08.03.550647. bioRxiv. 2023. Update in: Elife. 2024 Apr 15;12:RP91678. doi: 10.7554/eLife.91678. PMID: 37577495 Free PMC article. Updated. Preprint.
References
-
- Anderson DJ, Le Moigne R, Djakovic S, Kumar B, Rice J, Wong S, Wang J, Yao B, Valle E, Kiss von Soly S, Madriaga A, Soriano F, Menon MK, Wu ZY, Kampmann M, Chen Y, Weissman JS, Aftab BT, Yakes FM, Shawver L, Zhou HJ, Wustrow D, Rolfe M. Targeting the aaa atpase p97 as an approach to treat cancer through disruption of protein homeostasis. Cancer Cell. 2015;28:653–665. doi: 10.1016/j.ccell.2015.10.002. - DOI - PMC - PubMed
-
- Bartelt A, Widenmaier SB, Schlein C, Johann K, Goncalves RLS, Eguchi K, Fischer AW, Parlakgül G, Snyder NA, Nguyen TB, Bruns OT, Franke D, Bawendi MG, Lynes MD, Leiria LO, Tseng YH, Inouye KE, Arruda AP, Hotamisligil GS. Brown adipose tissue thermogenic adaptation requires Nrf1-mediated proteasomal activity. Nature Medicine. 2018;24:292–303. doi: 10.1038/nm.4481. - DOI - PMC - PubMed
-
- Bianchi G, Oliva L, Cascio P, Pengo N, Fontana F, Cerruti F, Orsi A, Pasqualetto E, Mezghrani A, Calbi V, Palladini G, Giuliani N, Anderson KC, Sitia R, Cenci S. The proteasome load versus capacity balance determines apoptotic sensitivity of multiple myeloma cells to proteasome inhibition. Blood. 2009;113:3040–3049. doi: 10.1182/blood-2008-08-172734. - DOI - PubMed
-
- Britton M, Lucas MM, Downey SL, Screen M, Pletnev AA, Verdoes M, Tokhunts RA, Amir O, Goddard AL, Pelphrey PM, Wright DL, Overkleeft HS, Kisselev AF. Selective inhibitor of proteasome’s caspase-like sites sensitizes cells to specific inhibition of chymotrypsin-like sites. Chemistry & Biology. 2009;16:1278–1289. doi: 10.1016/j.chembiol.2009.11.015. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

