mARC1 in MASLD: Modulation of lipid accumulation in human hepatocytes and adipocytes
- PMID: 38619429
- PMCID: PMC11019821
- DOI: 10.1097/HC9.0000000000000365
mARC1 in MASLD: Modulation of lipid accumulation in human hepatocytes and adipocytes
Abstract
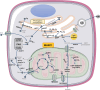
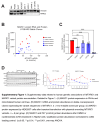
Background: Mutations in the gene MTARC1 (mitochondrial amidoxime-reducing component 1) protect carriers from metabolic dysfunction-associated steatohepatitis (MASH) and cirrhosis. MTARC1 encodes the mARC1 enzyme, which is localized to the mitochondria and has no known MASH-relevant molecular function. Our studies aimed to expand on the published human genetic mARC1 data and to observe the molecular effects of mARC1 modulation in preclinical MASH models.
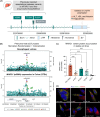
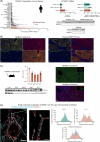
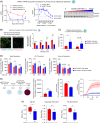
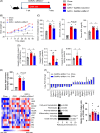
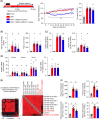
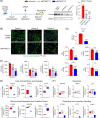
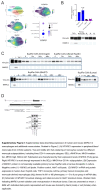
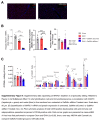
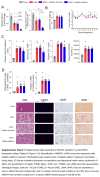
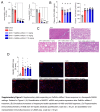
Methods and results: We identified a novel human structural variant deletion in MTARC1, which is associated with various biomarkers of liver health, including alanine aminotransferase levels. Phenome-wide Mendelian Randomization analyses additionally identified novel putatively causal associations between MTARC1 expression, and esophageal varices and cardiorespiratory traits. We observed that protective MTARC1 variants decreased protein accumulation in in vitro overexpression systems and used genetic tools to study mARC1 depletion in relevant human and mouse systems. Hepatocyte mARC1 knockdown in murine MASH models reduced body weight, liver steatosis, oxidative stress, cell death, and fibrogenesis markers. mARC1 siRNA treatment and overexpression modulated lipid accumulation and cell death consistently in primary human hepatocytes, hepatocyte cell lines, and primary human adipocytes. mARC1 depletion affected the accumulation of distinct lipid species and the expression of inflammatory and mitochondrial pathway genes/proteins in both in vitro and in vivo models.
Conclusions: Depleting hepatocyte mARC1 improved metabolic dysfunction-associated steatotic liver disease-related outcomes. Given the functional role of mARC1 in human adipocyte lipid accumulation, systemic targeting of mARC1 should be considered when designing mARC1 therapies. Our data point to plasma lipid biomarkers predictive of mARC1 abundance, such as Ceramide 22:1. We propose future areas of study to describe the precise molecular function of mARC1, including lipid trafficking and subcellular location within or around the mitochondria and endoplasmic reticulum.
Copyright © 2024 The Author(s). Published by Wolters Kluwer Health, Inc. on behalf of the American Association for the Study of Liver Diseases.
Conflict of interest statement
All authors are presently employed by or have received research funding from Boehringer Ingelheim. Dmitriy Drichel consults for Boehringer Ingelheim and Merck. Martin Giera owns stock in Gilead and GlaxoSmithKline.
Figures




References
-
- Rinella ME, Lazarus JV, Ratziu V, Francque SM, Sanyal AJ, Kanwal F, et al. . A multi-society Delphi consensus statement on new fatty liver disease nomenclature. J Hepatol. 2023;78:1966–1986. - PubMed
-
- Sanyal AJ, Harrison SA, Ratziu V, Abdelmalek MF, Diehl AM, Caldwell S, et al. . The natural history of advanced fibrosis due to nonalcoholic steatohepatitis: Data from the simtuzumab trials. Hepatology. 2019;70:1913–1927. - PubMed
-
- Younossi ZM, Wong G, Anstee QM, Henry L. The global burden of liver disease. Clin Gastroenterol Hepatol. 2023;21:1978–1991. - PubMed
-
- Plessis J du, Pelt J van, Korf H, Mathieu C, Schueren B van der, Lannoo M, et al. . Association of adipose tissue inflammation with histologic severity of nonalcoholic fatty liver disease. Gastroenterology. 2015;149:635–648.e14. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

