Iron regulates the quiescence of naive CD4 T cells by controlling mitochondria and cellular metabolism
- PMID: 38621136
- PMCID: PMC11047099
- DOI: 10.1073/pnas.2318420121
Iron regulates the quiescence of naive CD4 T cells by controlling mitochondria and cellular metabolism
Abstract
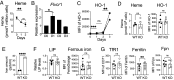
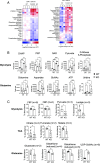
In response to an immune challenge, naive T cells undergo a transition from a quiescent to an activated state acquiring the effector function. Concurrently, these T cells reprogram cellular metabolism, which is regulated by iron. We and others have shown that iron homeostasis controls proliferation and mitochondrial function, but the underlying mechanisms are poorly understood. Given that iron derived from heme makes up a large portion of the cellular iron pool, we investigated iron homeostasis in T cells using mice with a T cell-specific deletion of the heme exporter, FLVCR1 [referred to as knockout (KO)]. Our finding revealed that maintaining heme and iron homeostasis is essential to keep naive T cells in a quiescent state. KO naive CD4 T cells exhibited an iron-overloaded phenotype, with increased spontaneous proliferation and hyperactive mitochondria. This was evidenced by reduced IL-7R and IL-15R levels but increased CD5 and Nur77 expression. Upon activation, however, KO CD4 T cells have defects in proliferation, IL-2 production, and mitochondrial functions. Iron-overloaded CD4 T cells failed to induce mitochondrial iron and exhibited more fragmented mitochondria after activation, making them susceptible to ferroptosis. Iron overload also led to inefficient glycolysis and glutaminolysis but heightened activity in the hexosamine biosynthetic pathway. Overall, these findings highlight the essential role of iron in controlling mitochondrial function and cellular metabolism in naive CD4 T cells, critical for maintaining their quiescent state.
Keywords: heme; iron; mitochondria; tonic signaling.
Conflict of interest statement
Competing interests statement:In the past 3 y, C.A.L. has consulted for Astellas Pharmaceuticals, Odyssey Therapeutics, Third Rock Ventures, and T-Knife Therapeutics, and is an inventor on patents pertaining to Kras regulated metabolic pathways, redox control pathways in pancreatic cancer, and targeting the GOT1-ME1 pathway as a therapeutic approach (US Patent No: 2015126580-A1, 05/07/2015; US Patent No: 20190136238, 05/09/2019; International Patent No: WO2013177426-A2, 04/23/2015).
Figures




References
-
- Stefanova I., Dorfman J. R., Germain R. N., Self-recognition promotes the foreign antigen sensitivity of naive T lymphocytes. Nature 420, 429–434 (2002). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

