Bone Proteomics Method Optimization for Forensic Investigations
- PMID: 38621258
- PMCID: PMC11077585
- DOI: 10.1021/acs.jproteome.4c00151
Bone Proteomics Method Optimization for Forensic Investigations
Abstract
The application of proteomic analysis to forensic skeletal remains has gained significant interest in improving biological and chronological estimations in medico-legal investigations. To enhance the applicability of these analyses to forensic casework, it is crucial to maximize throughput and proteome recovery while minimizing interoperator variability and laboratory-induced post-translational protein modifications (PTMs). This work compared different workflows for extracting, purifying, and analyzing bone proteins using liquid chromatography with tandem mass spectrometry (LC-MS)/MS including an in-StageTip protocol previously optimized for forensic applications and two protocols using novel suspension-trap technology (S-Trap) and different lysis solutions. This study also compared data-dependent acquisition (DDA) with data-independent acquisition (DIA). By testing all of the workflows on 30 human cortical tibiae samples, S-Trap workflows resulted in increased proteome recovery with both lysis solutions tested and in decreased levels of induced deamidations, and the DIA mode resulted in greater sensitivity and window of identification for the identification of lower-abundance proteins, especially when open-source software was utilized for data processing in both modes. The newly developed S-Trap protocol is, therefore, suitable for forensic bone proteomic workflows and, particularly when paired with DIA mode, can offer improved proteomic outcomes and increased reproducibility, showcasing its potential in forensic proteomics and contributing to achieving standardization in bone proteomic analyses for forensic applications.
Keywords: acquisition modes; bone proteomics; forensic science; mass spectrometry; protein extraction.
Conflict of interest statement
The authors declare no competing financial interest.
Figures



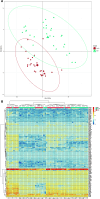

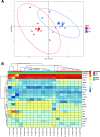
References
-
- Choi K. M.; Zissler A.; Kim E.; Ehrenfellner B.; Cho E.; Lee S. in.; Steinbacher P.; Yun K. N.; Shin J. H.; Kim J. Y.; Stoiber W.; Chung H.; Monticelli F. C.; Kim J. Y.; Pittner S. Postmortem Proteomics to Discover Biomarkers for Forensic PMI Estimation. Int. J. Legal Med. 2019, 133 (3), 899–908. 10.1007/s00414-019-02011-6. - DOI - PMC - PubMed
-
- Brockbals L.; Garrett-Rickman S.; Fu S.; Ueland M.; McNevin D.; Padula M. P. Estimating the Time of Human Decomposition Based on Skeletal Muscle Biopsy Samples Utilizing an Untargeted LC–MS/MS-Based Proteomics Approach. Anal. Bioanal. Chem. 2023, 415 (22), 5487–5498. 10.1007/s00216-023-04822-4. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

