Optimisation and evaluation of viral genomic sequencing of SARS-CoV-2 rapid diagnostic tests: a laboratory and cohort-based study
- PMID: 38621394
- PMCID: PMC11322816
- DOI: 10.1016/S2666-5247(23)00399-3
Optimisation and evaluation of viral genomic sequencing of SARS-CoV-2 rapid diagnostic tests: a laboratory and cohort-based study
Abstract
Background: Sequencing of SARS-CoV-2 from rapid diagnostic tests (RDTs) can bolster viral genomic surveillance efforts; however, approaches to maximise and standardise pathogen genome recovery from RDTs remain underdeveloped. We aimed to systematically optimise the elution of genetic material from RDT components and to evaluate the efficacy of RDT sequencing for outbreak investigation.
Methods: In this laboratory and cohort-based study we seeded RDTs with inactivated SARS-CoV-2 to optimise the elution of genomic material from RDT lateral flow strips. We measured the effect of changes in buffer type, time in buffer, and rotation on PCR cycle threshold (Ct) value. We recruited individuals older than 18 years residing in the greater Boston area, MA, USA, from July 18 to Nov 5, 2022, via email advertising to students and staff at Harvard University, MA, USA, and via broad social media advertising. All individuals recruited were within 5 days of a positive diagnostic test for SARS-CoV-2; no other relevant exclusion criteria were applied. Each individual completed two RDTs and one PCR swab. On Dec 29, 2022, we also collected RDTs from a convenience sample of individuals who were positive for SARS-CoV-2 and associated with an outbreak at a senior housing facility in MA, USA. We extracted all returned PCR swabs and RDT components (ie, swab, strip, or buffer); samples with a Ct of less than 40 were subject to amplicon sequencing. We compared the efficacy of elution and sequencing across RDT brands and components and used RDT-derived sequences to infer transmission links within the outbreak at the senior housing facility. We conducted metagenomic sequencing of negative RDTs from symptomatic individuals living in the senior housing facility.
Findings: Neither elution duration of greater than 10 min nor rotation during elution impacted viral titres. Elution in Buffer AVL (Ct=31·4) and Tris-EDTA Buffer (Ct=30·8) were equivalent (p=0·34); AVL outperformed elution in lysis buffer and 50% lysis buffer (Ct=40·0, p=0·0029 for both) as well as Universal Viral Transport Medium (Ct=36·7, p=0·079). Performance of RDT strips was poorer than that of matched PCR swabs (mean Ct difference 10·2 [SD 4·3], p<0·0001); however, RDT swabs performed similarly to PCR swabs (mean Ct difference 4·1 [5·2], p=0·055). No RDT brand significantly outperformed another. Across sample types, viral load predicted the viral genome assembly length. We assembled greater than 80% complete genomes from 12 of 17 RDT-derived swabs, three of 18 strips, and four of 11 residual buffers. We generated outbreak-associated SARS-CoV-2 genomes using both amplicon and metagenomic sequencing and identified multiple introductions of the virus that resulted in downstream transmission.
Interpretation: RDT-derived swabs are a reasonable alternative to PCR swabs for viral genomic surveillance and outbreak investigation. RDT-derived lateral flow strips yield accurate, but significantly fewer, viral reads than matched PCR swabs. Metagenomic sequencing of negative RDTs can identify viruses that might underlie patient symptoms.
Funding: The National Science Foundation, the Hertz Foundation, the National Institute of General Medical Sciences, Harvard Medical School, the Howard Hughes Medical Institute, the US Centers for Disease Control and Prevention, the Broad Institute and the National Institute of Allergy and Infectious Diseases.
Copyright © 2023 The Author(s). Published by Elsevier Ltd.. All rights reserved.
Conflict of interest statement
Declaration of interests PCS is a cofounder of, shareholder in, and advisor to Sherlock Biosciences; a board member of and shareholder in the Danaher Corporation; a cofounder of and shareholder in Delve Bio; and a shareholder in NextGenJane and TruGenomix. MS is a cofounder of, shareholder in, and advisor to Rhinostics; a shareholder of and advisor to Nexus Laboratories; and a consultant for Vectis Consulting. All other authors declare no competing interests.
Figures
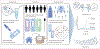


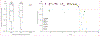
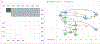
References
-
- DNA Pipelines R&D, Farr B, Rajan D, et al. COVID-19 ARTIC v4.1 Illumina library construction and sequencing protocol - tailed method V.2 March 31, 2022. 10.17504/protocols.io.j8nlk4b36g5r/v2 (accessed Oct 12, 2023). - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

