Sm-like protein Rof inhibits transcription termination factor ρ by binding site obstruction and conformational insulation
- PMID: 38622114
- PMCID: PMC11018626
- DOI: 10.1038/s41467-024-47439-6
Sm-like protein Rof inhibits transcription termination factor ρ by binding site obstruction and conformational insulation
Abstract
Transcription termination factor ρ is a hexameric, RNA-dependent NTPase that can adopt active closed-ring and inactive open-ring conformations. The Sm-like protein Rof, a homolog of the RNA chaperone Hfq, inhibits ρ-dependent termination in vivo but recapitulation of this activity in vitro has proven difficult and the precise mode of Rof action is presently unknown. Here, our cryo-EM structures of ρ-Rof and ρ-RNA complexes show that Rof undergoes pronounced conformational changes to bind ρ at the protomer interfaces, undercutting ρ conformational dynamics associated with ring closure and occluding extended primary RNA-binding sites that are also part of interfaces between ρ and RNA polymerase. Consistently, Rof impedes ρ ring closure, ρ-RNA interactions and ρ association with transcription elongation complexes. Structure-guided mutagenesis coupled with functional assays confirms that the observed ρ-Rof interface is required for Rof-mediated inhibition of cell growth and ρ-termination in vitro. Bioinformatic analyses reveal that Rof is restricted to Pseudomonadota and that the ρ-Rof interface is conserved. Genomic contexts of rof differ between Enterobacteriaceae and Vibrionaceae, suggesting distinct modes of Rof regulation. We hypothesize that Rof and other cellular anti-terminators silence ρ under diverse, but yet to be identified, stress conditions when unrestrained transcription termination by ρ may be detrimental.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


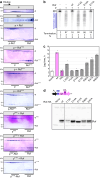
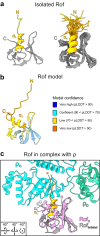




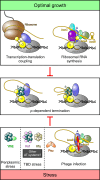
Update of
-
Sm-like protein Rof inhibits transcription termination factor ρ by binding site obstruction and conformational insulation.bioRxiv [Preprint]. 2023 Aug 31:2023.08.30.555460. doi: 10.1101/2023.08.30.555460. bioRxiv. 2023. Update in: Nat Commun. 2024 Apr 15;15(1):3186. doi: 10.1038/s41467-024-47439-6. PMID: 37693585 Free PMC article. Updated. Preprint.
Similar articles
-
A widely conserved protein Rof inhibits transcription termination factor Rho and promotes Salmonella virulence program.Nat Commun. 2024 Apr 15;15(1):3187. doi: 10.1038/s41467-024-47438-7. Nat Commun. 2024. PMID: 38622116 Free PMC article.
-
Sm-like protein Rof inhibits transcription termination factor ρ by binding site obstruction and conformational insulation.bioRxiv [Preprint]. 2023 Aug 31:2023.08.30.555460. doi: 10.1101/2023.08.30.555460. bioRxiv. 2023. Update in: Nat Commun. 2024 Apr 15;15(1):3186. doi: 10.1038/s41467-024-47439-6. PMID: 37693585 Free PMC article. Updated. Preprint.
-
The Sm-like RNA chaperone Hfq mediates transcription antitermination at Rho-dependent terminators.EMBO J. 2011 Jun 14;30(14):2805-16. doi: 10.1038/emboj.2011.192. EMBO J. 2011. PMID: 21673658 Free PMC article.
-
Rho-dependent terminators and transcription termination.Microbiology (Reading). 2006 Sep;152(Pt 9):2515-2528. doi: 10.1099/mic.0.28982-0. Microbiology (Reading). 2006. PMID: 16946247 Review.
-
Keeping up to speed with the transcription termination factor Rho motor.Transcription. 2010 Sep-Oct;1(2):70-5. doi: 10.4161/trns.1.2.12232. Transcription. 2010. PMID: 21326894 Free PMC article. Review.
Cited by
-
Simultaneous ligand binding to intact and partially formed ATP binding sites in the hexameric termination factor Rho.bioRxiv [Preprint]. 2025 Jul 24:2025.07.22.665897. doi: 10.1101/2025.07.22.665897. bioRxiv. 2025. PMID: 40777331 Free PMC article. Preprint.
-
A widely conserved protein Rof inhibits transcription termination factor Rho and promotes Salmonella virulence program.Nat Commun. 2024 Apr 15;15(1):3187. doi: 10.1038/s41467-024-47438-7. Nat Commun. 2024. PMID: 38622116 Free PMC article.
-
The Psu protein of phage satellite P4 inhibits transcription termination factor ρ by forced hyper-oligomerization.Nat Commun. 2025 Jan 9;16(1):550. doi: 10.1038/s41467-025-55897-9. Nat Commun. 2025. PMID: 39788982 Free PMC article.
-
c-di-GMP does not bind H-NS, nor inhibits H-NS binding DNA.Nat Commun. 2025 Jun 12;16(1):5287. doi: 10.1038/s41467-025-60689-2. Nat Commun. 2025. PMID: 40506437 Free PMC article. No abstract available.
-
Nucleotide-induced hyper-oligomerization inactivates transcription termination factor ρ.Nat Commun. 2025 Feb 15;16(1):1653. doi: 10.1038/s41467-025-56824-8. Nat Commun. 2025. PMID: 39952913 Free PMC article.
References
-
- Sunday, N., Svetlov, D. & Artsimovitch, I. Rho termination factor: one ring to bind them all. in RNA Polymerases as Molecular Motors: On the Road (eds. Landick, R., Strick, T. & Wang, J.) 100–131 (Royal Society of Chemistry, 2021).
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

