Single cell analysis unveils B cell-dominated immune subtypes in HNSCC for enhanced prognostic and therapeutic stratification
- PMID: 38622125
- PMCID: PMC11018606
- DOI: 10.1038/s41368-024-00292-1
Single cell analysis unveils B cell-dominated immune subtypes in HNSCC for enhanced prognostic and therapeutic stratification
Abstract
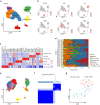
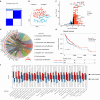
Head and neck squamous cell carcinoma (HNSCC) is characterized by high recurrence or distant metastases rate and the prognosis is challenging. There is mounting evidence that tumor-infiltrating B cells (TIL-Bs) have a crucial, synergistic role in tumor control. However, little is known about the role TIL-Bs play in immune microenvironment and the way TIL-Bs affect the outcome of immune checkpoint blockade. Using single-cell RNA sequencing (scRNA-seq) data from the Gene Expression Omnibus (GEO) database, the study identified distinct gene expression patterns in TIL-Bs. HNSCC samples were categorized into TIL-Bs inhibition and TIL-Bs activation groups using unsupervised clustering. This classification was further validated with TCGA HNSCC data, correlating with patient prognosis, immune cell infiltration, and response to immunotherapy. We found that the B cells activation group exhibited a better prognosis, higher immune cell infiltration, and distinct immune checkpoint levels, including elevated PD-L1. A prognostic model was also developed and validated, highlighting four genes as potential biomarkers for predicting survival outcomes in HNSCC patients. Overall, this study provides a foundational approach for B cells-based tumor classification in HNSCC, offering insights into targeted treatment and immunotherapy strategies.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

