Microenvironmental reorganization in brain tumors following radiotherapy and recurrence revealed by hyperplexed immunofluorescence imaging
- PMID: 38622132
- PMCID: PMC11018859
- DOI: 10.1038/s41467-024-47185-9
Microenvironmental reorganization in brain tumors following radiotherapy and recurrence revealed by hyperplexed immunofluorescence imaging
Abstract
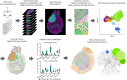
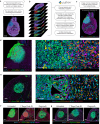
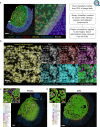
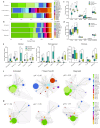
The tumor microenvironment plays a crucial role in determining response to treatment. This involves a series of interconnected changes in the cellular landscape, spatial organization, and extracellular matrix composition. However, assessing these alterations simultaneously is challenging from a spatial perspective, due to the limitations of current high-dimensional imaging techniques and the extent of intratumoral heterogeneity over large lesion areas. In this study, we introduce a spatial proteomic workflow termed Hyperplexed Immunofluorescence Imaging (HIFI) that overcomes these limitations. HIFI allows for the simultaneous analysis of > 45 markers in fragile tissue sections at high magnification, using a cost-effective high-throughput workflow. We integrate HIFI with machine learning feature detection, graph-based network analysis, and cluster-based neighborhood analysis to analyze the microenvironment response to radiation therapy in a preclinical model of glioblastoma, and compare this response to a mouse model of breast-to-brain metastasis. Here we show that glioblastomas undergo extensive spatial reorganization of immune cell populations and structural architecture in response to treatment, while brain metastases show no comparable reorganization. Our integrated spatial analyses reveal highly divergent responses to radiation therapy between brain tumor models, despite equivalent radiotherapy benefit.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Akkari, L. et al. Dynamic changes in glioma macrophage populations after radiotherapy reveal CSF-1R inhibition as a strategy to overcome resistance. Sci. Transl. Med.1210.1126/scitranslmed.aaw7843 (2020). - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

