Loss of SREBP-1c ameliorates iron-induced liver fibrosis by decreasing lipocalin-2
- PMID: 38622198
- PMCID: PMC11058876
- DOI: 10.1038/s12276-024-01213-2
Loss of SREBP-1c ameliorates iron-induced liver fibrosis by decreasing lipocalin-2
Abstract
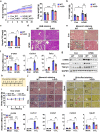
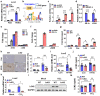
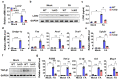
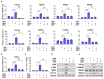
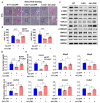
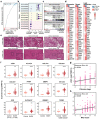
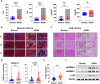
Sterol regulatory element-binding protein (SREBP)-1c is involved in cellular lipid homeostasis and cholesterol biosynthesis and is highly increased in nonalcoholic steatohepatitis (NASH). However, the molecular mechanism by which SREBP-1c regulates hepatic stellate cells (HSCs) activation in NASH animal models and patients have not been fully elucidated. In this study, we examined the role of SREBP-1c in NASH and the regulation of LCN2 gene expression. Wild-type and SREBP-1c knockout (1cKO) mice were fed a high-fat/high-sucrose diet, treated with carbon tetrachloride (CCl4), and subjected to lipocalin-2 (LCN2) overexpression. The role of LCN2 in NASH progression was assessed using mouse primary hepatocytes, Kupffer cells, and HSCs. LCN2 expression was examined in samples from normal patients and those with NASH. LCN2 gene expression and secretion increased in CCl4-induced liver fibrosis mice model, and SREBP-1c regulated LCN2 gene transcription. Moreover, treatment with holo-LCN2 stimulated intracellular iron accumulation and fibrosis-related gene expression in mouse primary HSCs, but these effects were not observed in 1cKO HSCs, indicating that SREBP-1c-induced LCN2 expression and secretion could stimulate HSCs activation through iron accumulation. Furthermore, LCN2 expression was strongly correlated with inflammation and fibrosis in patients with NASH. Our findings indicate that SREBP-1c regulates Lcn2 gene expression, contributing to diet-induced NASH. Reduced Lcn2 expression in 1cKO mice protects against NASH development. Therefore, the activation of Lcn2 by SREBP-1c establishes a new connection between iron and lipid metabolism, affecting inflammation and HSCs activation. These findings may lead to new therapeutic strategies for NASH.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

