Age-specific nasal epithelial responses to SARS-CoV-2 infection
- PMID: 38622380
- PMCID: PMC11087271
- DOI: 10.1038/s41564-024-01658-1
Age-specific nasal epithelial responses to SARS-CoV-2 infection
Erratum in
-
Author Correction: Age-specific nasal epithelial responses to SARS-CoV-2 infection.Nat Microbiol. 2024 Nov;9(11):3076. doi: 10.1038/s41564-024-01757-z. Nat Microbiol. 2024. PMID: 38890492 Free PMC article. No abstract available.
Abstract
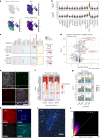
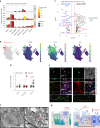
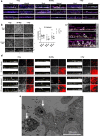
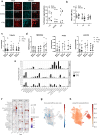
Children infected with SARS-CoV-2 rarely progress to respiratory failure. However, the risk of mortality in infected people over 85 years of age remains high. Here we investigate differences in the cellular landscape and function of paediatric (<12 years), adult (30-50 years) and older adult (>70 years) ex vivo cultured nasal epithelial cells in response to infection with SARS-CoV-2. We show that cell tropism of SARS-CoV-2, and expression of ACE2 and TMPRSS2 in nasal epithelial cell subtypes, differ between age groups. While ciliated cells are viral replication centres across all age groups, a distinct goblet inflammatory subtype emerges in infected paediatric cultures and shows high expression of interferon-stimulated genes and incomplete viral replication. In contrast, older adult cultures infected with SARS-CoV-2 show a proportional increase in basaloid-like cells, which facilitate viral spread and are associated with altered epithelial repair pathways. We confirm age-specific induction of these cell types by integrating data from in vivo COVID-19 studies and validate that our in vitro model recapitulates early epithelial responses to SARS-CoV-2 infection.
© 2024. The Author(s).
Conflict of interest statement
In the past three years, S.A.T. has received remuneration for Scientific Advisory Board Membership from Sanofi, GlaxoSmithKline, Foresite Labs and Qiagen. S.A.T. is a co-founder and holds equity in Transition Bio and Ensocell. Starting 8 January 2024, S.A.T. has been a part-time employee of GlaxoSmithKline. All other authors declare no competing interests.
Figures











References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

