High-risk Escherichia coli clones that cause neonatal meningitis and association with recrudescent infection
- PMID: 38622998
- PMCID: PMC11021048
- DOI: 10.7554/eLife.91853
High-risk Escherichia coli clones that cause neonatal meningitis and association with recrudescent infection
Abstract
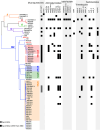
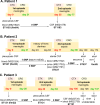
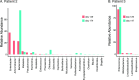
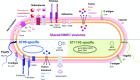
Neonatal meningitis is a devastating disease associated with high mortality and neurological sequelae. Escherichia coli is the second most common cause of neonatal meningitis in full-term infants (herein NMEC) and the most common cause of meningitis in preterm neonates. Here, we investigated the genomic relatedness of a collection of 58 NMEC isolates spanning 1974-2020 and isolated from seven different geographic regions. We show NMEC are comprised of diverse sequence types (STs), with ST95 (34.5%) and ST1193 (15.5%) the most common. No single virulence gene profile was conserved in all isolates; however, genes encoding fimbrial adhesins, iron acquisition systems, the K1 capsule, and O antigen types O18, O75, and O2 were most prevalent. Antibiotic resistance genes occurred infrequently in our collection. We also monitored the infection dynamics in three patients that suffered recrudescent invasive infection caused by the original infecting isolate despite appropriate antibiotic treatment based on antibiogram profile and resistance genotype. These patients exhibited severe gut dysbiosis. In one patient, the causative NMEC isolate was also detected in the fecal flora at the time of the second infection episode and after treatment. Thus, although antibiotics are the standard of care for NMEC treatment, our data suggest that failure to eliminate the causative NMEC that resides intestinally can lead to the existence of a refractory reservoir that may seed recrudescent infection.
Keywords: E. coli; genomics; gut dysbiosis; infectious disease; microbiology; neonatal meningitis; recurrent infection; uropathogenic Escherichia coli.
© 2023, Nhu et al.
Conflict of interest statement
NN, MP, SH, KP, LA, BF, SA, TM, PH, SB, SS, HB, PT, AB, BW, AI, MS No competing interests declared
Figures




Update of
References
-
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. Journal of Computational Biology. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. - DOI - PMC - PubMed
-
- Bidet P, Mahjoub-Messai F, Blanco J, Blanco J, Dehem M, Aujard Y, Bingen E, Bonacorsi S. Combined multilocus sequence typing and O serogrouping distinguishes Escherichia coli subtypes associated with infant urosepsis and/or meningitis. The Journal of Infectious Diseases. 2007;196:297–303. doi: 10.1086/518897. - DOI - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

