Vascular endothelial cells-derived exosomes synergize with curcumin to prevent osteoporosis development
- PMID: 38623340
- PMCID: PMC11016789
- DOI: 10.1016/j.isci.2024.109608
Vascular endothelial cells-derived exosomes synergize with curcumin to prevent osteoporosis development
Abstract
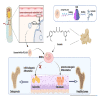
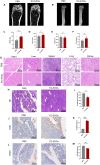
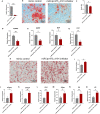
Osteoporosis has gradually become a major public health problem. Further elucidation of the pathophysiological mechanisms that induce osteoporosis and identification of more effective therapeutic targets will have important clinical significance. Experiments in vitro on bone marrow stem cells (BMSCs) subjected to osteogenic and adipogenic differentiation and in vivo on surgical bilateral ovariectomy (OVX) mouse models revealed that exosomes of vascular endothelial cells (EC-EXOs) can promote osteogenic differentiation of BMSCs and inhibit BMSC adipogenic differentiation through miR-3p-975_4191. Both miR-3p-975_4191 and curcumin can target tumor necrosis factor (TNF) and act synergistically to regulate BMSCs fate differentiation and delay the progression of osteoporosis. Our findings suggest that EC-EXOs may exert a synergistic effect with curcumin in reversing the progression of osteoporosis by targeting TNF via miR-3p-975_4191. Our study may provide therapeutic options and potential therapeutic targets for osteoporosis and thus has important clinical implications.
Keywords: Cell biology; Orthopedics; Stem cells research.
© 2024 The Authors.
Conflict of interest statement
The authors declare no competing interests.
Figures








Similar articles
-
Mechanism of EC-EXOs-Derived THBS3 Targeting CD47 to Regulate BMSCs Differentiation to Ameliorate Bone Loss.Cell Prolif. 2025 Jun 13:e70066. doi: 10.1111/cpr.70066. Online ahead of print. Cell Prolif. 2025. PMID: 40514348
-
MiR-210 promotes bone formation in ovariectomized rats by regulating osteogenic/adipogenic differentiation of bone marrow mesenchymal stem cells through downregulation of EPHA2.J Orthop Surg Res. 2023 Oct 30;18(1):811. doi: 10.1186/s13018-023-04213-6. J Orthop Surg Res. 2023. PMID: 37904187 Free PMC article.
-
ERK-estrogen receptor α signaling plays a role in the process of bone marrow mesenchymal stem cell-derived exosomes protecting against ovariectomy-induced bone loss.J Orthop Surg Res. 2023 Mar 27;18(1):250. doi: 10.1186/s13018-023-03660-5. J Orthop Surg Res. 2023. PMID: 36973789 Free PMC article.
-
Mechanism of vascular endothelial cell-derived exosomes modified with vascular endothelial growth factor in steroid-induced femoral head necrosis.Biomed Mater. 2023 Feb 16;18(2). doi: 10.1088/1748-605X/acb412. Biomed Mater. 2023. PMID: 36794758
-
Melatonin promotes bone marrow mesenchymal stem cell osteogenic differentiation and prevents osteoporosis development through modulating circ_0003865 that sponges miR-3653-3p.Stem Cell Res Ther. 2021 Feb 25;12(1):150. doi: 10.1186/s13287-021-02224-w. Stem Cell Res Ther. 2021. PMID: 33632317 Free PMC article.
Cited by
-
Comprehensive systematic review and meta-analysis on the therapeutic efficacy of curcumin in osteoporosis: unveiling mechanisms and preclinical evidence.Front Nutr. 2025 May 21;12:1590256. doi: 10.3389/fnut.2025.1590256. eCollection 2025. Front Nutr. 2025. PMID: 40469677 Free PMC article. Review.
-
Exosome‑mediated crosstalk between the cardiovascular and musculoskeletal systems: Mechanisms and therapeutic potential (Review).Int J Mol Med. 2025 Sep;56(3):129. doi: 10.3892/ijmm.2025.5570. Epub 2025 Jun 27. Int J Mol Med. 2025. PMID: 40576128 Free PMC article. Review.
-
Efficient and high-density immobilization of animal cells by a microfiber with both swelling and cell adhesion properties and its application to exosome production.Biotechnol Lett. 2025 Apr 12;47(2):40. doi: 10.1007/s10529-025-03585-5. Biotechnol Lett. 2025. PMID: 40220210 Free PMC article.
-
Endothelial cell-derived exosomes trigger a positive feedback loop in osteogenesis-angiogenesis coupling via up-regulating zinc finger and BTB domain containing 16 in bone marrow mesenchymal stem cell.J Nanobiotechnology. 2024 Nov 19;22(1):721. doi: 10.1186/s12951-024-03002-5. J Nanobiotechnology. 2024. PMID: 39563357 Free PMC article.
References
-
- Maes C., Kobayashi T., Selig M.K., Torrekens S., Roth S.I., Mackem S., Carmeliet G., Kronenberg H.M. Osteoblast precursors, but not mature osteoblasts, move into developing and fractured bones along with invading blood vessels. Dev. Cell. 2010;19:329–344. doi: 10.1016/j.devcel.2010.07.010. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

