Genomic analysis of human brain metastases treated with stereotactic radiosurgery reveals unique signature based on treatment failure
- PMID: 38623341
- PMCID: PMC11016778
- DOI: 10.1016/j.isci.2024.109601
Genomic analysis of human brain metastases treated with stereotactic radiosurgery reveals unique signature based on treatment failure
Abstract
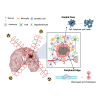
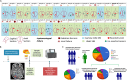
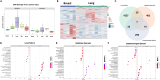
Stereotactic radiosurgery (SRS) has been shown to be efficacious for the treatment of limited brain metastasis (BM); however, the effects of SRS on human brain metastases have yet to be studied. We performed genomic analysis on resected brain metastases from patients whose resected lesion was previously treated with SRS. Our analyses demonstrated for the first time that patients possess a distinct genomic signature based on type of treatment failure including local failure, leptomeningeal spread, and radio-necrosis. Examination of the center and peripheral edge of the tumors treated with SRS indicated differential DNA damage distribution and an enrichment for tumor suppressor mutations and DNA damage repair pathways along the peripheral edge. Furthermore, the two clinical modalities used to deliver SRS, LINAC and GK, demonstrated differential effects on the tumor landscape even between controlled primary sites. Our study provides, in human, biological evidence of differential effects of SRS across BM's.
Keywords: Cancer; Cancer systems biology; Genomic analysis; Genomics.
© 2024 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Brown P.D., Ballman K.V., Cerhan J.H., Anderson S.K., Carrero X.W., Whitton A.C., Greenspoon J., Parney I.F., Laack N.N.I., Ashman J.B., et al. Postoperative stereotactic radiosurgery compared with whole brain radiotherapy for resected metastatic brain disease (NCCTG N107C/CEC·3): a multicentre, randomised, controlled, phase 3 trial. Lancet Oncol. 2017;18:1049–1060. doi: 10.1016/s1470-2045(17)30441-2. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

