The estrogen signaling pathway reprograms prostate cancer cell metabolism and supports proliferation and disease progression
- PMID: 38625747
- PMCID: PMC11142735
- DOI: 10.1172/JCI170809
The estrogen signaling pathway reprograms prostate cancer cell metabolism and supports proliferation and disease progression
Abstract
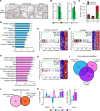
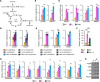
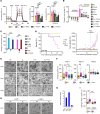
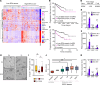
Just like the androgen receptor (AR), the estrogen receptor α (ERα) is expressed in the prostate and is thought to influence prostate cancer (PCa) biology. Yet the incomplete understanding of ERα functions in PCa hinders our ability to fully comprehend its clinical relevance and restricts the repurposing of estrogen-targeted therapies for the treatment of this disease. Using 2 human PCa tissue microarray cohorts, we first demonstrate that nuclear ERα expression was heterogeneous among patients, being detected in only half of the tumors. Positive nuclear ERα levels were correlated with disease recurrence, progression to metastatic PCa, and patient survival. Using in vitro and in vivo models of the normal prostate and PCa, bulk and single-cell RNA-Seq analyses revealed that estrogens partially mimicked the androgen transcriptional response and activated specific biological pathways linked to proliferation and metabolism. Bioenergetic flux assays and metabolomics confirmed the regulation of cancer metabolism by estrogens, supporting proliferation. Using cancer cell lines and patient-derived organoids, selective estrogen receptor modulators, a pure anti-estrogen, and genetic approaches impaired cancer cell proliferation and growth in an ERα-dependent manner. Overall, our study revealed that, when expressed, ERα functionally reprogrammed PCa metabolism, was associated with disease progression, and could be targeted for therapeutic purposes.
Keywords: Endocrinology; Glucose metabolism; Oncology; Prostate cancer; Sex hormones.
Conflict of interest statement
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

